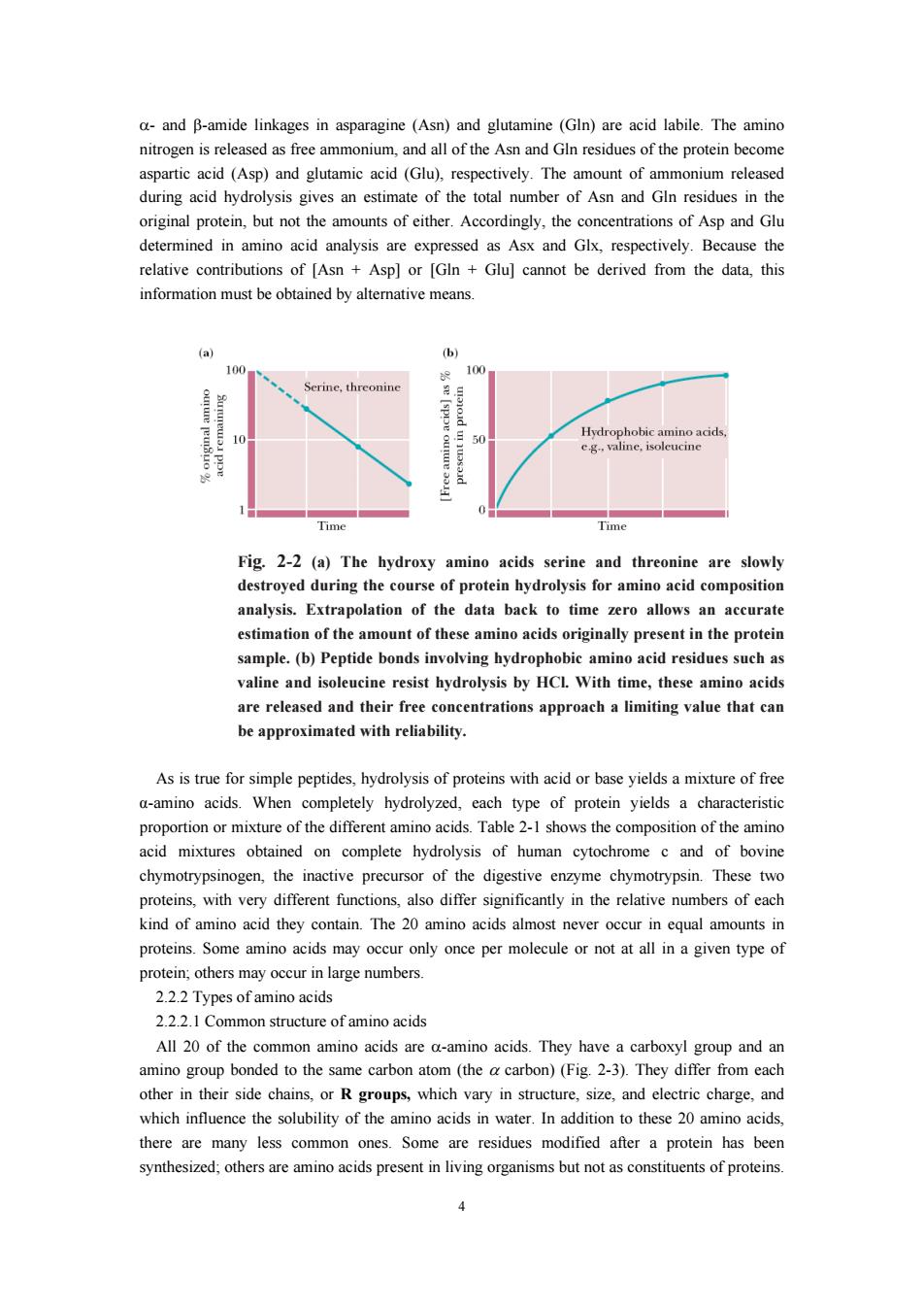
a-and B-amide linkages in asparagine (Asn)and glutamine (GIn)are acid labile.The amino nitrogen is released as free ammonium,and al of the Asn and G residues of the protein become (Asp)and glutamic acid (Glu) respectively The amount of rovem o thoa her of and Cto original protein,but not the amounts of either.Accordingly,the concentrations of Asp and Glu determined in amino acid analysis are expressed as Asx and Glx,respectively.Because the relative contributions of IAsn Aspl or [GIn Glul cannot be derived from the data.this information must be obtained by altemative means Time Time Fig.2-2(a)The hydroxy amino acids serine and threonine are slov destroyed during the course of protein hydrolysis for amino acid composition analysis.Extrapolation of the data back to time zero allows an accurate estimation of the amount of these amino acids originally present in the protein sample.(b)Peptide bonds involving hvdrophobic amino acid residues such as valine and isoleucine resist hydrolysis by HCl With time,these amino acids are released and their free concentrations approach a limiting value that can be approximated with reliability. As is true for simple peptides,hydrolysis of proteins with acid or base yields a mixture of free a-amino acids.When completely hydrolyzed,each type of protein yields a characteristic portion rmixture of the diffe nt amir tion of the amino ned on complete hydrolysis of human cytochrome c and of bovine chymotrypsinogen,the inactive precursor of the digestive enzyme chymotrypsin.These two proteins,with very different functions,also differ significantly in the relative numbers of each kind of amino acid they contain.The 20 amino acids almost never occur in equal amounts in only once per molecule or not at all in a given type of 2.22Types ofamino acids 222.1 Common structure of amino acids All 20 of the common amino acids are a-amino acids.They have a carboxyl group and an amino group bonded to the same carbor atom(the carbon)(Fig 2-3).They differ from each other in their side chains. or Rgroups which vary in structure,size,and eletric charge.and which influence the solubility of the amino acids in water.In addition to these 20 amino acids there are many less common ones.Some are residues modified after a protein has been synthesized:others are amino acids present in living organisms but not as constituents of proteins. 4
4 α- and β-amide linkages in asparagine (Asn) and glutamine (Gln) are acid labile. The amino nitrogen is released as free ammonium, and all of the Asn and Gln residues of the protein become aspartic acid (Asp) and glutamic acid (Glu), respectively. The amount of ammonium released during acid hydrolysis gives an estimate of the total number of Asn and Gln residues in the original protein, but not the amounts of either. Accordingly, the concentrations of Asp and Glu determined in amino acid analysis are expressed as Asx and Glx, respectively. Because the relative contributions of [Asn + Asp] or [Gln + Glu] cannot be derived from the data, this information must be obtained by alternative means. Fig. 2-2 (a) The hydroxy amino acids serine and threonine are slowly destroyed during the course of protein hydrolysis for amino acid composition analysis. Extrapolation of the data back to time zero allows an accurate estimation of the amount of these amino acids originally present in the protein sample. (b) Peptide bonds involving hydrophobic amino acid residues such as valine and isoleucine resist hydrolysis by HCl. With time, these amino acids are released and their free concentrations approach a limiting value that can be approximated with reliability. As is true for simple peptides, hydrolysis of proteins with acid or base yields a mixture of free α-amino acids. When completely hydrolyzed, each type of protein yields a characteristic proportion or mixture of the different amino acids. Table 2-1 shows the composition of the amino acid mixtures obtained on complete hydrolysis of human cytochrome c and of bovine chymotrypsinogen, the inactive precursor of the digestive enzyme chymotrypsin. These two proteins, with very different functions, also differ significantly in the relative numbers of each kind of amino acid they contain. The 20 amino acids almost never occur in equal amounts in proteins. Some amino acids may occur only once per molecule or not at all in a given type of protein; others may occur in large numbers. 2.2.2 Types of amino acids 2.2.2.1 Common structure of amino acids All 20 of the common amino acids are α-amino acids. They have a carboxyl group and an amino group bonded to the same carbon atom (the α carbon) (Fig. 2-3). They differ from each other in their side chains, or R groups, which vary in structure, size, and electric charge, and which influence the solubility of the amino acids in water. In addition to these 20 amino acids, there are many less common ones. Some are residues modified after a protein has been synthesized; others are amino acids present in living organisms but not as constituents of proteins
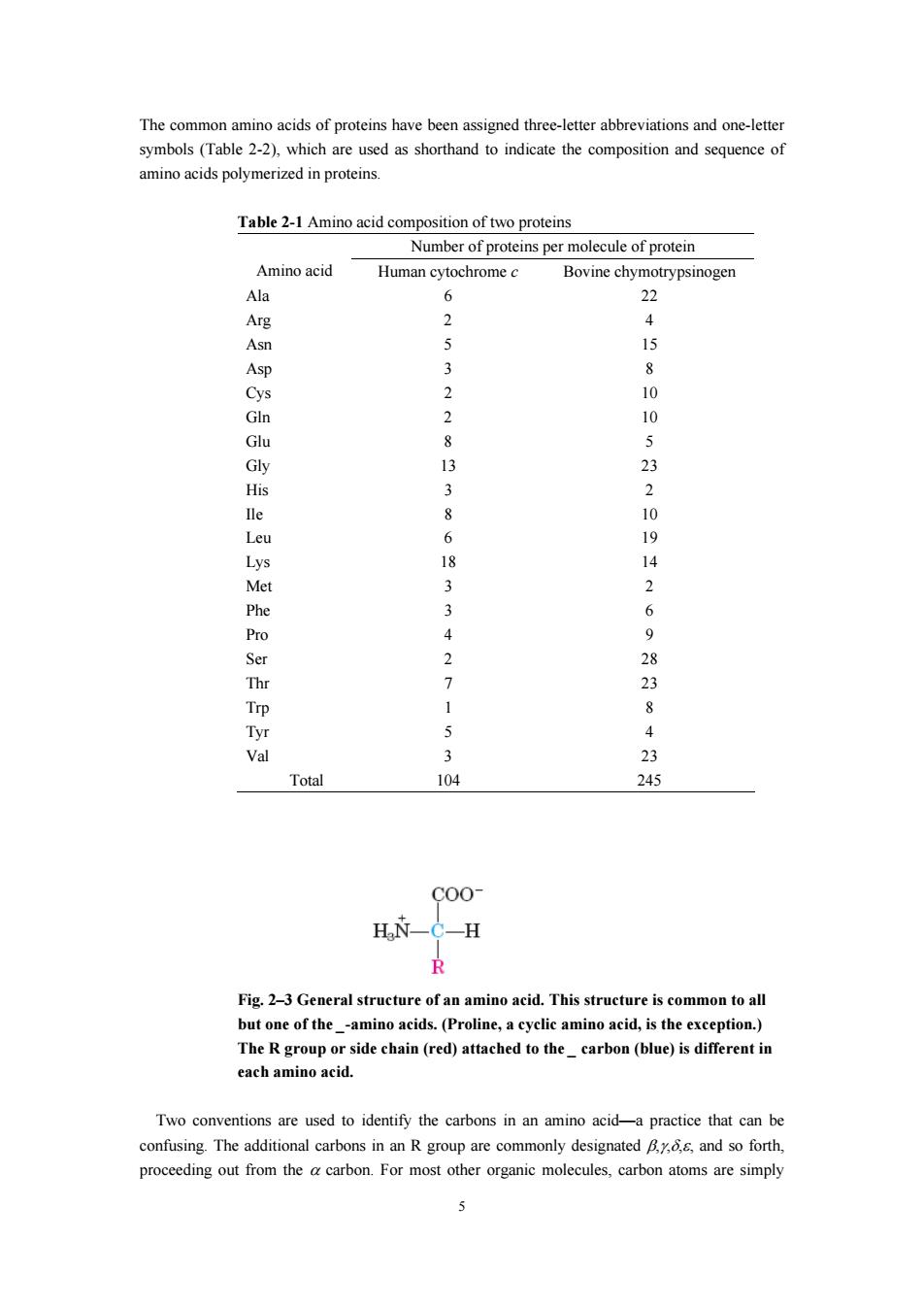
The common amino acids of proteins have been assigned three-letter abbreviations and one-letter which are shornd o indicste the composiion and se of rized in proteins Table 2-1 Amino acid composition of two proteins Number of proteins per molecule of protein Amino acid Human cytochromec Bovine chyn 0 2 4 Asn 5 15 10 Glu 8 Gly 13 His 3 2 8 6P8 14 3 Phe Pro 4 9 2 Tyr Val 3 23 Total 104 245 C00 H C-H Fig.2-3 General structure of an amino acid.This structure is common to all butone of theamino acids.(Proline,acyclic amino acid,is the exception.) The R groupo ide (red)attached to thecarbon(blue)is each amino acic Two conventions are used to identify the carbons in an amino acid-a practice that can be confusing.The additional carbons in an R group are commony designatedand so forth proceeding out from thecabon.For most organic molecules atoms are simply
5 The common amino acids of proteins have been assigned three-letter abbreviations and one-letter symbols (Table 2-2), which are used as shorthand to indicate the composition and sequence of amino acids polymerized in proteins. Table 2-1 Amino acid composition of two proteins Number of proteins per molecule of protein Amino acid Human cytochrome c Bovine chymotrypsinogen Ala 6 22 Arg 2 4 Asn 5 15 Asp 3 8 Cys 2 10 Gln 2 10 Glu 8 5 Gly 13 23 His 3 2 Ile 8 10 Leu 6 19 Lys 18 14 Met 3 2 Phe 3 6 Pro 4 9 Ser 2 28 Thr 7 23 Trp 1 8 Tyr 5 4 Val 3 23 Total 104 245 Fig. 2–3 General structure of an amino acid. This structure is common to all but one of the _-amino acids. (Proline, a cyclic amino acid, is the exception.) The R group or side chain (red) attached to the _ carbon (blue) is different in each amino acid. Two conventions are used to identify the carbons in an amino acid—a practice that can be confusing. The additional carbons in an R group are commonly designated β,γ,δ,ε, and so forth, proceeding out from the α carbon. For most other organic molecules, carbon atoms are simply
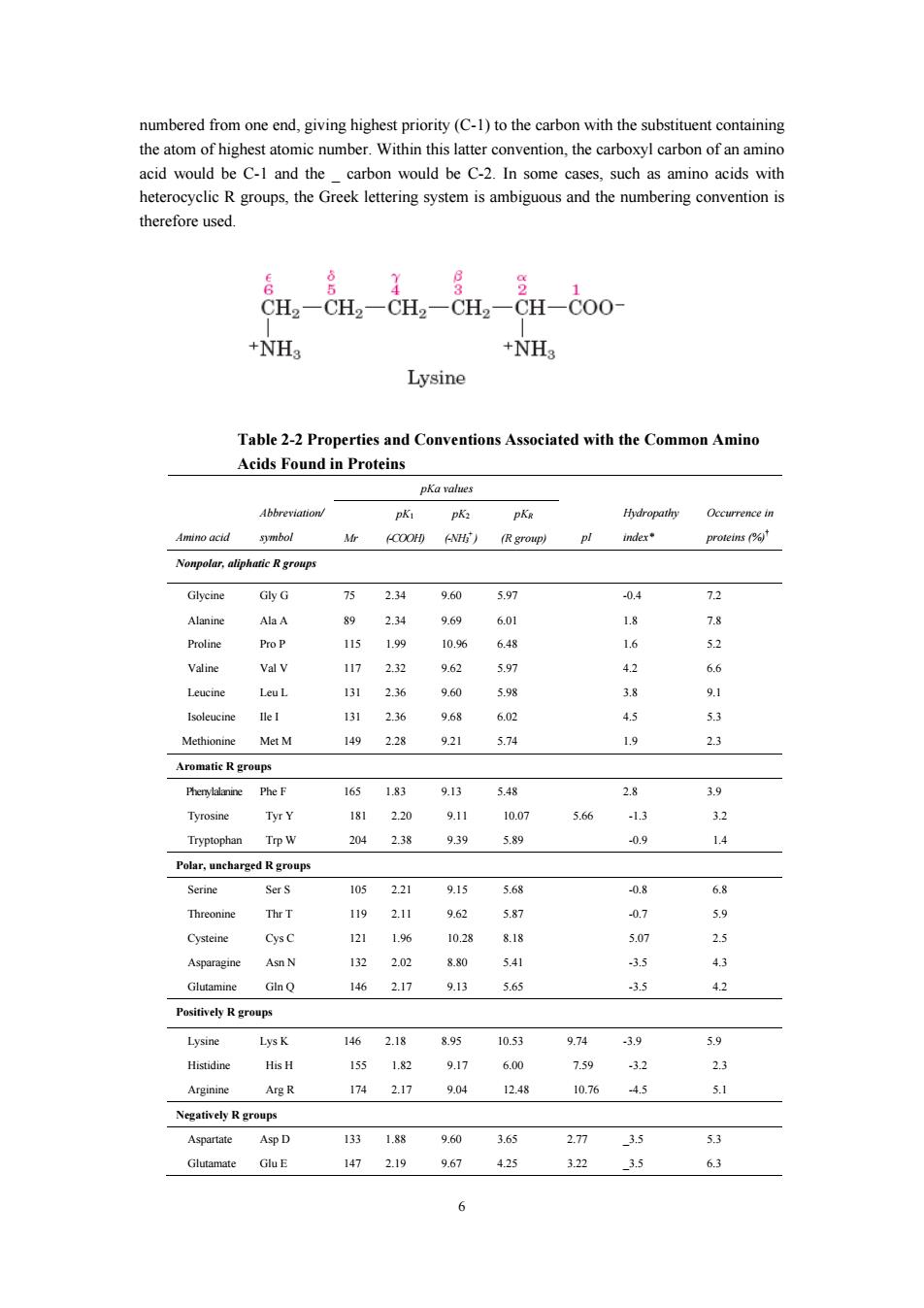
numbered from one end,giving highest priority(C-1)to the carbon with the substituent containing the atom of highest atomic number.Within this atter of an amino acid would be C-1 and the carbor would be C-.In some cases such as amino acids with heterocyclic R groups,the Greek lettering system is ambiguous and the numbering convention i therefore used. CHCH-CH.CH CH-Coo +NH +NH Lysine Table 2-2 Properties s and Conventions Associated with the Common Amind Acids Found in Proteins pKa values Occurrence in imino acid (COO (R gromp) inder Nonpolar,aliphatic R groups 234 04 72 AlaA 89 2.34 969 601 Pro P 115 1.99 10.96 68 Valine Val v 117 232 6 42 6 Leut 13 2.36 96 38 131 236 50 1492.28 921 5.74 23 Aromatic R groups 165 1g3 9.13 38 3.9 230 911 566 32 238 939 5.9 14 Polar,uncharged Rgroups Serine SerS 105 221 915 568 -0.8 68 1192.11 9.62 07 14 25 5 43 1462.17 9.13 5.65 3.5 42 Powi价e小R groups Lysine 2.18 895 10.53 974 5.9 Histidine His H 155 1.82 91 6.00 -32 23 Arginine A段R 1742.17 9.04 1248 10.76 .4.5 5.1 A甲I 13 18器 3 53 147 2.19 9.67 425 322 3.5 6.3 6
6 numbered from one end, giving highest priority (C-1) to the carbon with the substituent containing the atom of highest atomic number. Within this latter convention, the carboxyl carbon of an amino acid would be C-1 and the _ carbon would be C-2. In some cases, such as amino acids with heterocyclic R groups, the Greek lettering system is ambiguous and the numbering convention is therefore used. Table 2-2 Properties and Conventions Associated with the Common Amino Acids Found in Proteins pKa values Amino acid Abbreviation/ symbol Mr pK1 (-COOH) pK2 (-NH3 + ) pKR (R group) pI Hydropathy index* Occurrence in proteins (%)† Nonpolar, aliphatic R groups Glycine Gly G 75 2.34 9.60 5.97 -0.4 7.2 Alanine Ala A 89 2.34 9.69 6.01 1.8 7.8 Proline Pro P 115 1.99 10.96 6.48 1.6 5.2 Valine Val V 117 2.32 9.62 5.97 4.2 6.6 Leucine Leu L 131 2.36 9.60 5.98 3.8 9.1 Isoleucine Ile I 131 2.36 9.68 6.02 4.5 5.3 Methionine Met M 149 2.28 9.21 5.74 1.9 2.3 Aromatic R groups Phenylalanine Phe F 165 1.83 9.13 5.48 2.8 3.9 Tyrosine Tyr Y 181 2.20 9.11 10.07 5.66 -1.3 3.2 Tryptophan Trp W 204 2.38 9.39 5.89 -0.9 1.4 Polar, uncharged R groups Serine Ser S 105 2.21 9.15 5.68 -0.8 6.8 Threonine Thr T 119 2.11 9.62 5.87 -0.7 5.9 Cysteine Cys C 121 1.96 10.28 8.18 5.07 2.5 Asparagine Asn N 132 2.02 8.80 5.41 -3.5 4.3 Glutamine Gln Q 146 2.17 9.13 5.65 -3.5 4.2 Positively R groups Lysine Lys K 146 2.18 8.95 10.53 9.74 -3.9 5.9 Histidine His H 155 1.82 9.17 6.00 7.59 -3.2 2.3 Arginine Arg R 174 2.17 9.04 12.48 10.76 -4.5 5.1 Negatively R groups Aspartate Asp D 133 1.88 9.60 3.65 2.77 _3.5 5.3 Glutamate Glu E 147 2.19 9.67 4.25 3.22 _3.5 6.3
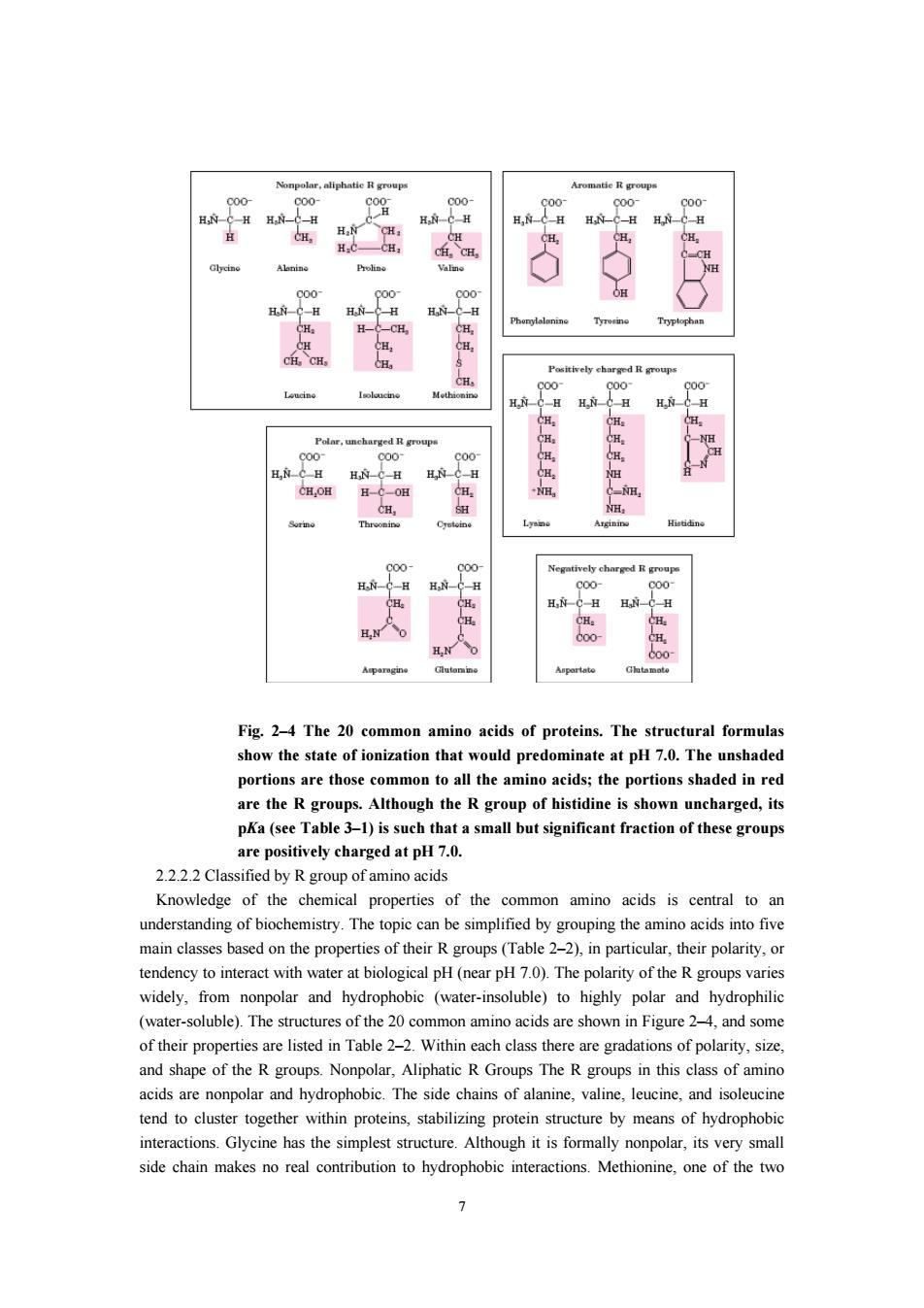
d度 H Fig.2-4 The 20 common amino acids of proteins.The structural formulas show the state of ionization that would predominate at pH 7.0.The unshaded ortions are those common to all the aminoaid wn uncharged,its pKa(see Table 3-1)is such that a small but significant fraction of these groups are positively charged at pH 7.0. 2.22.2 Classified by R group of amino acids Knowledge of the chemical properties of the common amino acids is central to an their polarity tendency to interact with water at biological pH(near pH7.0).The polarity of the R groups varies widely,from nonpolar and hydrophobic (water-insoluble)to highly polar and hydrophilic (water-soluble).The structures of the 20 common amino acids are shown in Figure 2-4,and some of their properties are listed in Table 2-2.Within each class there are gradations of polarity.size and shape of the Rgroups.Nonpolar,Aliphatic R Groups The R groups in this lass of amino cidsare nonpoar and hydrophobic.The side chains, tend to cluster together within proteins,stabilizing protein structure by means of hydrophobic interactions.Glycine has the simplest structure.Although it is formally nonpolar,its very small side chain makes no real contribution to hydrophobic interactions.Methionine,one of the two 7
7 Fig. 2–4 The 20 common amino acids of proteins. The structural formulas show the state of ionization that would predominate at pH 7.0. The unshaded portions are those common to all the amino acids; the portions shaded in red are the R groups. Although the R group of histidine is shown uncharged, its pKa (see Table 3–1) is such that a small but significant fraction of these groups are positively charged at pH 7.0. 2.2.2.2 Classified by R group of amino acids Knowledge of the chemical properties of the common amino acids is central to an understanding of biochemistry. The topic can be simplified by grouping the amino acids into five main classes based on the properties of their R groups (Table 2–2), in particular, their polarity, or tendency to interact with water at biological pH (near pH 7.0). The polarity of the R groups varies widely, from nonpolar and hydrophobic (water-insoluble) to highly polar and hydrophilic (water-soluble). The structures of the 20 common amino acids are shown in Figure 2–4, and some of their properties are listed in Table 2–2. Within each class there are gradations of polarity, size, and shape of the R groups. Nonpolar, Aliphatic R Groups The R groups in this class of amino acids are nonpolar and hydrophobic. The side chains of alanine, valine, leucine, and isoleucine tend to cluster together within proteins, stabilizing protein structure by means of hydrophobic interactions. Glycine has the simplest structure. Although it is formally nonpolar, its very small side chain makes no real contribution to hydrophobic interactions. Methionine, one of the two

sulfur-containing amino acids,has a nonpolar thioether group in its side chain.Proline has an residues is tion that reduces the of polypeptide regions containing proline. 2.2.3 Properties of amino acids 2.2.3.1 Optical rotation of amino acids Except for glyeine,all of the amino acids isolated from proteins have four different groups carbon atom.In such a case.the a-carbor is said to be thGrt sheirinnd th we constitute nonsuperimposable mirror image isomers,or enantiomers).Enantiomeric molecules display a special property called optical activity-the ability to rotate the plane of polarization of plane-polarized light.Clockwise rotation of incident light is referred to as dextrorotatory behavior and rotation is called levorotatory behavior.The magnitude and direction of the dep nd on the nature of the amino acid side chain. The temperatu wavelength of the light used in the measurement,the ionization state of the amino acid,and therefore the pH of the soution,can also affect optical rotation behavior.As shown in Table2-5 some protein-derived amino acids at a given pH are dextrorotatory and others are levorotatory ough all of them are of the figuration.The direction of ptical tation can be specified in the name by using a (+)for dextrorotatory compounds and a (-)for levorotatory compounds,as in L(+)-leucine Table 2-5 Specific rotations for some amino acids Amino acid Specific Rotation 2 Alanine +1.8 Arginine +125 Aspartic acid +5.0 Glutamic acid 4120 Histidine .385 Leucine -11.0 Lvsine +13.5 Methionine -10.0 Phenylalanine 34。 Serine Threonine -28.5 Tryptophan .337 Valine +5.6 The discoveries of optical activity and enantiomeric structures made it important to develo suitable nomenclature for chiral molecules.Two systems are in common use today:the so-called D.L system and the (R,S)system.In the D.L system of nomenclature,the(+)and(-)isomers of glyceraldehyde are denoted as D-glyceraldehyde and Lglyceraldehyde,respectively (Fig.2-5). 8
8 sulfur-containing amino acids, has a nonpolar thioether group in its side chain. Proline has an aliphatic side chain with a distinctive cyclic structure. The secondary amino (imino) group of proline residues is held in a rigid conformation that reduces the structural flexibility of polypeptide regions containing proline. 2.2.3 Properties of amino acids 2.2.3.1 Optical rotation of amino acids Except for glycine, all of the amino acids isolated from proteins have four different groups attached to the α-carbon atom. In such a case, the α-carbon is said to be asymmetric or chiral (from the Greek cheir, meaning “hand”), and the two possible configurations for the α-carbon constitute nonsuperimposable mirror image isomers, or enantiomers). Enantiomeric molecules display a special property called optical activity—the ability to rotate the plane of polarization of plane-polarized light. Clockwise rotation of incident light is referred to as dextrorotatory behavior, and counterclockwise rotation is called levorotatory behavior. The magnitude and direction of the optical rotation depend on the nature of the amino acid side chain. The temperature, the wavelength of the light used in the measurement, the ionization state of the amino acid, and therefore the pH of the solution, can also affect optical rotation behavior. As shown in Table 2-5, some protein-derived amino acids at a given pH are dextrorotatory and others are levorotatory, even though all of them are of the L configuration. The direction of optical rotation can be specified in the name by using a (+) for dextrorotatory compounds and a (-) for levorotatory compounds, as in L(+)-leucine. Table 2-5 Specific rotations for some amino acids Amino acid Specific Rotation [α] D 25, Alanine +1.8 Arginine +12.5 Aspartic acid +5.0 Glutamic acid +12.0 Histidine -38.5 Isoleucine +12.4 Leucine -11.0 Lysine +13.5 Methionine -10.0 Phenylalanine -34.5 Proline -86.2 Serine -7.5 Threonine -28.5 Tryptophan -33.7 Valine +5.6 The discoveries of optical activity and enantiomeric structures made it important to develop suitable nomenclature for chiral molecules. Two systems are in common use today: the so-called D,L system and the (R,S) system. In the D,L system of nomenclature, the (+) and (-) isomers of glyceraldehyde are denoted as D-glyceraldehyde and L-glyceraldehyde, respectively (Fig. 2-5)