
transcribed as the end product is often called an RNA gene or non-coding RNA gene Non-coding RNA genes include transfer RNA(tRNA)and ribosomal RNA (rRNA).small RNAs such as snoRNAs,microRNAs,SIRNAs and piRNAs and lastly long ncRNAs that include examples such as Xist,Evf,Air,CTN and PINK The number of neRNAs encoded within the genome is unknown,however recent transcriptomic and microarray studies suggest the existence of over 30,000 long ncRNAs and at leas s many small regulatory RNAs within then ouse genom alone.Since most of the newly identified ncRNAs have not been validated for their function,it is possible that the majority of them are meaningless(e.g.non-functional or truncated transcript). 2.4 The Structure of Chromosomes The term"chromosome"is used to refer to a nucleic acid molecule that is the repository of genetic information in a virus,a bacterium,a eukaryotic cell,or an organelle.It also refers to the densely colored bodies seen in the nuclei of dye-stained eukaryotic cells,as visualized using a light microscope. .41 Eukaryotic nucleosor 2.4.1.1 omponents.The eukaryotic cell cycle produces remarkable changes in the structure of chromosomes).In nondividing eukaryotic cells(in GO)and those in interphase(Gl,S,and G2),the chromosomal material,chromatin,is amorphous and appears to be randomly dispersed in certain parts of the nucleus.In the S phase of interphase the DNA in this amorphous state plicates,each chromosome s(called、 chr matids)h 1a5 other after replication is complete.The chromosomes become much more condensed during prophase of mitosis,taking the form of a speciesspecific number of well-defined pairs of sister chromatids. Chromatin consists of fibers containing protein and DNA in approximately equal masses,along with a small ar ount of RNA.The DNA in the chr atin is very tightly associated with proteins called histones,which package and order the DNA into structural units called nucleosomes (Figure 2.5).Histone core Also found many nonhistone proteins.some of which help maintain chromosome structure.others that regulate the expression of specific genes.Beginning with nucleosomes.eukaryotic chromosomal DNA is packaged into a succession of higher-order structures that ultimately yield the compact chrom some seen with the light .We turn to a description of this structure in eukaryotes and compare it with the packaging of DNA in bacterial cells
21 transcribed as the end product is often called an RNA gene or non-coding RNA gene. Non-coding RNA genes include transfer RNA (tRNA) and ribosomal RNA (rRNA), small RNAs such as snoRNAs, microRNAs, siRNAs and piRNAs and lastly long ncRNAs that include examples such as Xist, Evf, Air, CTN and PINK. The number of ncRNAs encoded within the genome is unknown, however recent transcriptomic and microarray studies suggest the existence of over 30,000 long ncRNAs and at least as many small regulatory RNAs within the mouse genome alone. Since most of the newly identified ncRNAs have not been validated for their function, it is possible that the majority of them are meaningless (e.g. non-functional or truncated transcript). 2.4 The Structure of Chromosomes The term “chromosome” is used to refer to a nucleic acid molecule that is the repository of genetic information in a virus, a bacterium, a eukaryotic cell, or an organelle. It also refers to the densely colored bodies seen in the nuclei of dye-stained eukaryotic cells, as visualized using a light microscope. 2.4.1 Eukaryotic nucleosomes 2.4.1.1components. The eukaryotic cell cycle produces remarkable changes in the structure of chromosomes). In nondividing eukaryotic cells (in G0) and those in interphase (G1, S, and G2), the chromosomal material, chromatin, is amorphous and appears to be randomly dispersed in certain parts of the nucleus. In the S phase of interphase the DNA in this amorphous state replicates, each chromosome producing two sister chromosomes (called sister chromatids) that remain associated with each other after replication is complete. The chromosomes become much more condensed during prophase of mitosis, taking the form of a speciesspecific number of well-defined pairs of sister chromatids. Chromatin consists of fibers containing protein and DNA in approximately equal masses, along with a small amount of RNA. The DNA in the chromatin is very tightly associated with proteins called histones, which package and order the DNA into structural units called nucleosomes (Figure 2.5). Histone core Also found many nonhistone proteins, some of which help maintain chromosome structure, others that regulate the expression of specific genes. Beginning with nucleosomes, eukaryotic chromosomal DNA is packaged into a succession of higher-order structures that ultimately yield the compact chromosome seen with the light microscope. We now turn to a description of this structure in eukaryotes and compare it with the packaging of DNA in bacterial cells
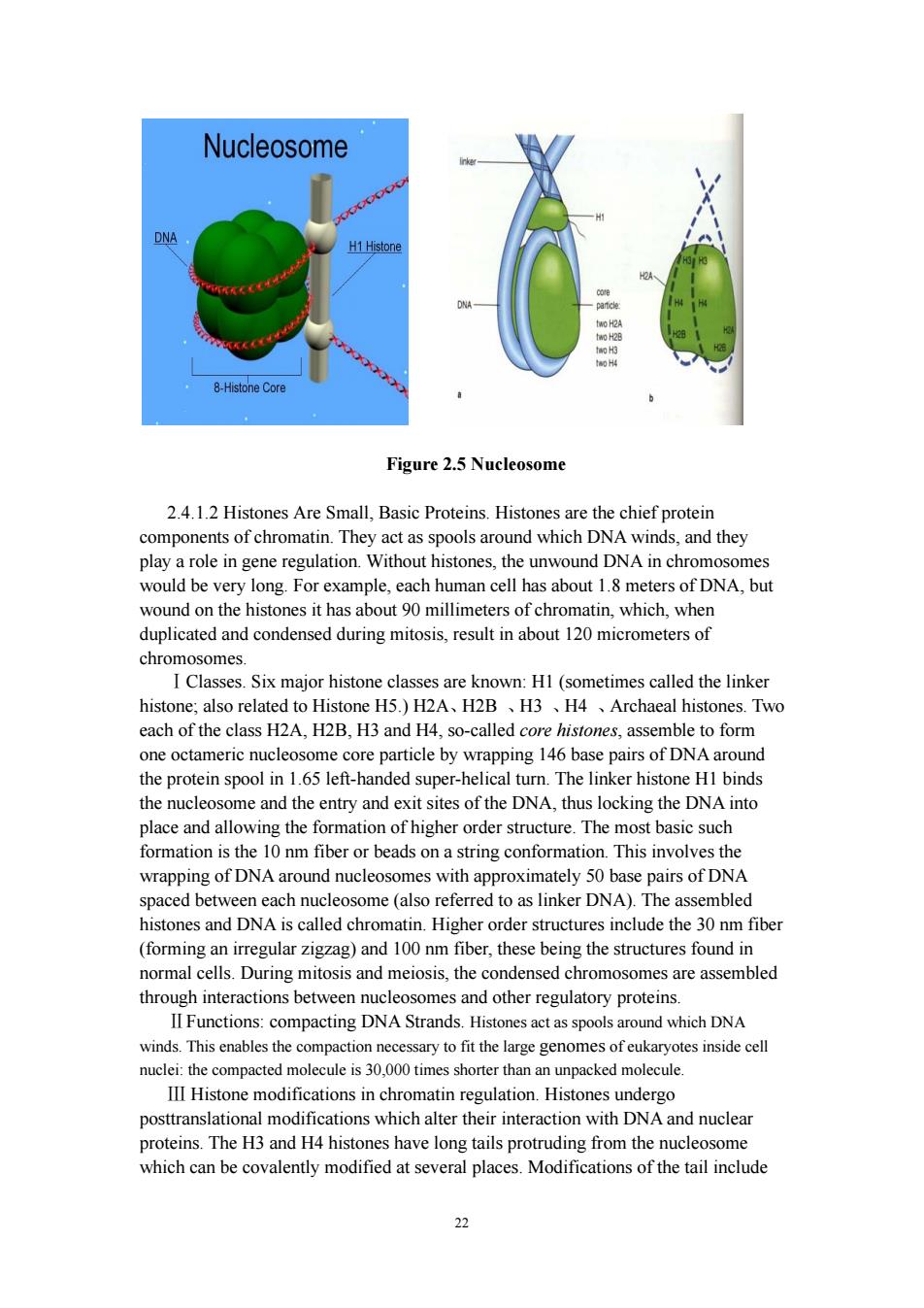
Nucleosome 。 8-Histone Core Figure 2.5 Nucleosome 2.4.1.2 Histones Are Small,Basic Proteins.Histones are the chief protein components of chromatin.They act as spools around which DNA winds,and they unwound DNA in chr wound on the histones it has about 90 millimeters of chromatin,which,when duplicated and condensed during mitosis,result in about 120 micrometers of chromosomes I Classe s Six maior histone classes are known:Hl (sometimes called the linker o Histone H5,)H2A、H2B、H3、H4 、Archaeal hist nes Two each of the class H2A,H2B,H3 and H4,so-called core histones,assemble to form one octameric nucleosome core particle by wrapping 146 base pairs of DNA around the protein spool in 1.65 left-handed super-helical turn.The linker histone HI binds the nucleosome and the entry and exit sites of the DNA,thus locking the DNA into lace and all wing the fo ofhigher orer structure.The m asic such formation is the 10 nm fiber or beads on a string conformation.This involves the wrapping of DNA around nucleosomes with approximately 50 base pairs of DNA spaced between each nucleosome(also referred to as linker DNA).The assembled histones and dna is called chromatin.higher order structures include the 30 nm fiber (forming an irregular zigzag)and 100 nm fiber,these being the structures found in normal cells.During mitosis and meiosis,the condensed chromosomes are assembled through interactions between nucleosomes and other regulatory proteins. II Functions:compacting DNA Strands.Histones act as spools around which DNA winds.This enables the compaction necessary to fit the large genomes of eukaryotes inside cell nucei:the comp acted molecule is 30 000 times shorter tha unpacked molecule. I Histone modifications in chromatin regulation.Histones undergo posttranslational modifications which alter their interaction with DNA and nuclear proteins.The H3 and H4 histones have long tails protruding from the nucleosome which can be covalently modified at several places.Modifications of the tail include
22 Figure 2.5 Nucleosome 2.4.1.2 Histones Are Small, Basic Proteins. Histones are the chief protein components of chromatin. They act as spools around which DNA winds, and they play a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA, but wound on the histones it has about 90 millimeters of chromatin, which, when duplicated and condensed during mitosis, result in about 120 micrometers of chromosomes. ⅠClasses. Six major histone classes are known: H1 (sometimes called the linker histone; also related to Histone H5.) H2A、H2B 、H3 、H4 、Archaeal histones. Two each of the class H2A, H2B, H3 and H4, so-called core histones, assemble to form one octameric nucleosome core particle by wrapping 146 base pairs of DNA around the protein spool in 1.65 left-handed super-helical turn. The linker histone H1 binds the nucleosome and the entry and exit sites of the DNA, thus locking the DNA into place and allowing the formation of higher order structure. The most basic such formation is the 10 nm fiber or beads on a string conformation. This involves the wrapping of DNA around nucleosomes with approximately 50 base pairs of DNA spaced between each nucleosome (also referred to as linker DNA). The assembled histones and DNA is called chromatin. Higher order structures include the 30 nm fiber (forming an irregular zigzag) and 100 nm fiber, these being the structures found in normal cells. During mitosis and meiosis, the condensed chromosomes are assembled through interactions between nucleosomes and other regulatory proteins. ⅡFunctions: compacting DNA Strands. Histones act as spools around which DNA winds. This enables the compaction necessary to fit the large genomes of eukaryotes inside cell nuclei: the compacted molecule is 30,000 times shorter than an unpacked molecule. Ⅲ Histone modifications in chromatin regulation. Histones undergo posttranslational modifications which alter their interaction with DNA and nuclear proteins. The H3 and H4 histones have long tails protruding from the nucleosome which can be covalently modified at several places. Modifications of the tail include

methylation.acetylation.phosphorylation.ubiquitination.sumovlation.citrullination and ADP-ribosylation.The core of the histones(H2A and H3)can also be modified. code".Histone modifications act in diverse biological processes such as gene regulation,DNA repair and chromosome condensation(mitosis). 2.4.2 Nucleosomes are packed into successively higher order structures .mfiber.Wrapping of DNA core compacts the DNA length about sevenfold.The overall compaction in a chromosome,however,is greater than 10.000-fold-ample evidence for even higher orders of structural organization.In chromosomes isolated by very gentle methods,nucleosome cores niedctre called the m fiber (Fire This es one molecule of histone HI per nucleoso me core.Organization into 30 nm fibers does not extend over the entire chromosome but is punctuated by regions bound by equence-specific (nonhistone)DNA binding proteins.The 30 nm structure also appears to depend on the transcriptional activity of the particular region of DNA Regions in which genes are being transcribed are apparently in a less-ordered state that contains little any histone HI.The 30 nm fiber,a second level of chro organization,provides an approximat tely 1-ol DNA. levels of folding are not yet understood,but it appears that certain regions of DNA associate with a nuclear scaffold(Figure 2.6).The scaffold-associated regions are separated by loops of DNA with perhaps 20 to 100 kbp.The DNA in a loop may ntain a set of related genes.For exan histone-coding genes gether in loops that are scaffold attachment sites(Figure 26).The scaffold itself appears to contain several proteins. notably large amounts of histone H1 (located in the interior of the fiber)and topoisomerase II.The presence of topoisomerase II further emphasizes the relationship between DNA underwinding and chromatin structure.Topoisomerase II is so important to the maintenance of ch omatin structure that inhibitors of this enzyme can kill rapidly Several drugs used incancer chemotherapy are topoisomerase II inhibitors that allow the enzyme to promote strand breakage but not the resealing of the breaks.Evidence exists for additional layers of organization in eukaryotic chromosomes,each dramatically enhancing the degree of compaction
23 methylation, acetylation, phosphorylation, ubiquitination, sumoylation, citrullination, and ADP-ribosylation. The core of the histones (H2A and H3) can also be modified. Combinations of modifications are thought to constitute a code, the so-called "histone code". Histone modifications act in diverse biological processes such as gene regulation, DNA repair and chromosome condensation (mitosis). 2.4.2 Nucleosomes are packed into successively higher order structures 2.4.2.1 30 nm fiber. Wrapping of DNA around a nucleosome core compacts the DNA length about sevenfold. The overall compaction in a chromosome, however, is greater than 10,000-fold—ample evidence for even higher orders of structural organization. In chromosomes isolated by very gentle methods, nucleosome cores appear to be organized into a structure called the 30 nm fiber (Figure 2.6). This packing requires one molecule of histone H1 per nucleosome core. Organization into 30 nm fibers does not extend over the entire chromosome but is punctuated by regions bound by equence-specific (nonhistone) DNA binding proteins. The 30 nm structure also appears to depend on the transcriptional activity of the particular region of DNA. Regions in which genes are being transcribed are apparently in a less-ordered state that contains little, if any, histone H1. The 30 nm fiber, a second level of chromatin organization, provides an approximately 100-fold compaction of the DNA. The higher levels of folding are not yet understood, but it appears that certain regions of DNA associate with a nuclear scaffold (Figure 2.6). The scaffold-associated regions are separated by loops of DNA with perhaps 20 to 100 kbp. The DNA in a loop may contain a set of related genes. For example, in Drosophila complete sets of histone-coding genes seem to cluster together in loops that are bounded by scaffold attachment sites (Figure 2.6). The scaffold itself appears to contain several proteins, notably large amounts of histone H1 (located in the interior of the fiber) and topoisomerase II. The presence of topoisomerase II further emphasizes the relationship between DNA underwinding and chromatin structure. Topoisomerase II is so important to the maintenance of chromatin structure that inhibitors of this enzyme can kill rapidly dividing cells. Several drugs used in cancer chemotherapy are topoisomerase II inhibitors that allow the enzyme to promote strand breakage but not the resealing of the breaks. Evidence exists for additional layers of organization in eukaryotic chromosomes, each dramatically enhancing the degree of compaction

30 nm Fiber Histone genes Figure 2.6 Nucleosomes are packed into successively higher order structures Higherorder chromatin structure probably varies from chromosome to chromosome,from one region to the next in a single chromosome,and from moment to moment in the life of a cell.No single model can adequately describe these structures.Nevertheless,the principle is clear:DNA cor paction in eukaryotic Beyond this the structure of chromatin is poorly understood,but it is classically suggested that the 30 nm fiber is arranged into loops along a central protein scaffold to form transcriptionally active euchromatin.Further compaction leads to transcriptionally inactive heterochromatin. 2.4.22EuCr atin and heterochro matin.Euchromatin is a lightly packed form of chromatin that is rich in gene concentration,and is often(but not always)unde active transcription.Unlike heterochromatin,it is found in both eukaryotes and prokaryotes. The structure of euchromatin is reminiscient of an unfolded set of beads along a string,where those beads represent nucleosomes.Nucleosomes consist of eight protei mas histones. ith approximately 146 base pairs of DNA wound around them:in euchromatin this wrapping is loose so that the raw DNA may be accessed.Each core histone possesses a'tail'structure which can vary in several ways;it is thought that these variations act as"master control switches"which determine the overall arrangement of the chromatin.In particular,it is believed that the presence of methylated lysine4on the histone tails cts as a general marker for euchromatin Euchromatin participates in the active transcription of DNA to mRNA products Heterochromatin is a tightly packed form of DNA.Its major characteristic is that transcription is limited.As such,it is a means to control gene expression,through regulation of the tra the protection of the integrity of chromosomes;all of these roles can be attributed to the dense packing of DNA,which makes it less accessible to protein factors that bind DNA or its associated factors. 24
24 Figure 2.6 Nucleosomes are packed into successively higher order structures. Higherorder chromatin structure probably varies from chromosome to chromosome, from one region to the next in a single chromosome, and from moment to moment in the life of a cell. No single model can adequately describe these structures. Nevertheless, the principle is clear: DNA compaction in eukaryotic chromosomes is likely to involve coils upon coils upon coils. Beyond this the structure of chromatin is poorly understood, but it is classically suggested that the 30 nm fiber is arranged into loops along a central protein scaffold to form transcriptionally active euchromatin. Further compaction leads to transcriptionally inactive heterochromatin. 2.4.2.2 Euchromatin and heterochromatin. Euchromatin is a lightly packed form of chromatin that is rich in gene concentration, and is often (but not always) under active transcription. Unlike heterochromatin, it is found in both eukaryotes and prokaryotes. The structure of euchromatin is reminiscient of an unfolded set of beads along a string, where those beads represent nucleosomes. Nucleosomes consist of eight proteins known as histones, with approximately 146 base pairs of DNA wound around them; in euchromatin this wrapping is loose so that the raw DNA may be accessed. Each core histone possesses a `tail' structure which can vary in several ways; it is thought that these variations act as "master control switches" which determine the overall arrangement of the chromatin. In particular, it is believed that the presence of methylated lysine 4 on the histone tails acts as a general marker for euchromatin. Euchromatin participates in the active transcription of DNA to mRNA products. Heterochromatin is a tightly packed form of DNA. Its major characteristic is that transcription is limited. As such, it is a means to control gene expression, through regulation of the transcription initiation. Heterochromatin is believed to serve several functions, from gene regulation to the protection of the integrity of chromosomes; all of these roles can be attributed to the dense packing of DNA, which makes it less accessible to protein factors that bind DNA or its associated factors

2.4.3 Chromosomes in prokaryotes The prok bacteria and arch typically have a single circular st bacteria have a single circula chromosome that can range in size from only 160,000 base pairs in the endosymbiotic bacteria Candidatus Carsonella ruddii,to 12,200,000 base pairs in the soil-dwelling bacteria Sorangium cellulosum.Spirochaetes of the genus Borrelia are a notable exception to this arrangement,with bacteria such as Borrelia burgdo eri,the cause of Lyme disease,containing a single linear chr 2.4.3.1 Structure in sequences.Prokaryotes chromosomes have less sequence-based structure than eukarvotes.Bacteria typically have a single point (the origin of replication)from which replication starts,while some archaea contain multiple replication origins.The genes in prokaryotes are often organised in operons. and do ain int s,unlike euka 2.4.3.2 DNA packaging Prokaryotes do not possess nuclei,instead their DNA is organized into a structure called the nucleoid.The nucleoid is a distinct structure and occupies a defined region of the bacterial cell.This structure is however dynamic and is maintained and remodeled by the actions of a range of histone-like proteins. which associate with the bacterial ch e In a the DNa in omosomes is even more organized,with the DNA packaged within structures similar to eukaryotic nucleosomes. Bacterial chromosomes tend to be tethered to the plasma membrane of the bacteria.In molecular biology application,this allows for its isolation from plasmid DNA by centrifugation of lysed bacteria and pelleting of the membranes(and the attached DNA). Prokaryotic chromosomes and plasmids are,like eukaryotic DNA,generally supercoiled.The DNA must first be released into its relaxed state for access for transcription,regulation,and replication. 2.5 Introduce to several important model organisms 2.5.1 What is model organism? A model organism is a species that is extensively studied to understand particular biological phenomena,with the expe nsight into the workings of other organ Inpcr explore potential causes and treatments for human disease when experimentation on humans would be unfeasible or unethical.This strategy is made possible by the common descent of all living organisms,and the conservation of metabolic and developmental pathways and genetic material over the course of evolution 2.5.2 Selecting a model organism Often,model organisms are chosen on the basis that they are amenable to experimental manipulation.This usually will include characteristics such as short life-cycle,techniques for genetic manipulation(inbred strains,stem cell lines,and transfection systems)and non-specialist living requirements.Sometimes,the genome
25 2.4.3 Chromosomes in prokaryotes The prokaryotes - bacteria and archaea - typically have a single circular chromosome, but many variations do exist. Most bacteria have a single circular chromosome that can range in size from only 160,000 base pairs in the endosymbiotic bacteria Candidatus Carsonella ruddii, to 12,200,000 base pairs in the soil-dwelling bacteria Sorangium cellulosum. Spirochaetes of the genus Borrelia are a notable exception to this arrangement, with bacteria such as Borrelia burgdorferi, the cause of Lyme disease, containing a single linear chromosome. 2.4.3.1 Structure in sequences. Prokaryotes chromosomes have less sequence-based structure than eukaryotes. Bacteria typically have a single point (the origin of replication) from which replication starts, while some archaea contain multiple replication origins. The genes in prokaryotes are often organised in operons, and do not contain introns, unlike eukaryotes. 2.4.3.2 DNA packaging. Prokaryotes do not possess nuclei, instead their DNA is organized into a structure called the nucleoid. The nucleoid is a distinct structure and occupies a defined region of the bacterial cell. This structure is however dynamic and is maintained and remodeled by the actions of a range of histone-like proteins, which associate with the bacterial chromosome. In archaea, the DNA in chromosomes is even more organized, with the DNA packaged within structures similar to eukaryotic nucleosomes. Bacterial chromosomes tend to be tethered to the plasma membrane of the bacteria. In molecular biology application, this allows for its isolation from plasmid DNA by centrifugation of lysed bacteria and pelleting of the membranes (and the attached DNA). Prokaryotic chromosomes and plasmids are, like eukaryotic DNA, generally supercoiled. The DNA must first be released into its relaxed state for access for transcription, regulation, and replication. 2.5 Introduce to several important model organisms 2.5.1 What is model organism? A model organism is a species that is extensively studied to understand particular biological phenomena, with the expectation that discoveries made in the organism model will provide insight into the workings of other organisms. In particular, model organisms are widely used to explore potential causes and treatments for human disease when experimentation on humans would be unfeasible or unethical. This strategy is made possible by the common descent of all living organisms, and the conservation of metabolic and developmental pathways and genetic material over the course of evolution. 2.5.2 Selecting a model organism Often, model organisms are chosen on the basis that they are amenable to experimental manipulation. This usually will include characteristics such as short life-cycle, techniques for genetic manipulation (inbred strains, stem cell lines, and transfection systems) and non-specialist living requirements. Sometimes, the genome