
2.1.2.2 Genomes and genetic variation.note that a genome does not capture the genetic genetic polymorphismof spe genome sequence in principle could the DNA of one cell from one individual.To learn what variations in genetic information underlie particular traits or diseases requires comparisons across individuals.This point explains the common usage of"genome"(which parallels a common usage of"gene")to refer not to the information in any particular DNA seque e.but toa whole family of sequences that share a biological context. Although this concept may seem counter intuitive,it is the same concept that says there is no particular shape that is the shape of a cheetah.Cheetahs vary,and so do the sequences of their genomes.Yet both the individual animals and their sequences share commonalities,so one can learn something about cheetahs and ch eetah-ness"from a single example of either 2.3 Characteristic of genome structure 2.3.1 Virus A virus (from the latin virus meaning "toxin"or "poison")is a sub-microscopic infectious agent that is unable to growor r repro tside a host cell.Each viral particle,or viri n,consists of genetic material,DNA or RNA, a protective protein coat called a capsid.The capsid shape varies from simple helical and icosahedral (polyhedral or near-spherical)forms,to more complex structures with tails or an envelope.Viruses infect cellular life forms and are grouped into animal. plant and bacterial types,according to the type of host infected. An enormou s can be seer ong viral pecies;as a group they contain more structural genomic diversity than the entire kingdoms of either plants,animals,or bacteria. 2.3.1.1 Size.Genome size in terms of the weight of nucleotides varies between species.The smallest genomes code for only four proteins and weigh about 10 Daltons.the larges eigh about sand hundred proten RNA viruses generally have smaller genome sizes than DNA viruses d due to a higher error-rate when replicating,resulting in a maximum upper size limit.Beyond this limit,errors in the genome when replicating render the virus useless or uncompetitive. To compensate for this,RNA viruses often have segmented genomes where the genome is split into smaller molecules,thus reducing the chance of error.In contrast. replication enzymes 2.3.12 Genome.A virus may employ either DNA or RNA as the nucleic acid Rarely do they contain both,however cytomegalovirus is an exception to this, possessing a DNA core with several mRNA segments.By far most viruses have RNA Plant viruse e single-stranded RNA and acte end to hav double-stranded DNA.Some virus species possess abnormal nucleotides,such as hydroxymethylcytosine instead of cytosine,as a normal part of their genome. Viral genomes may be circular,such as polyomaviruses,or linear,such as adenoviruses.The type of nucleic acid is irrelevant to the shape of the genome
11 2.1.2.2 Genomes and genetic variation. Note that a genome does not capture the genetic diversity or the genetic polymorphism of a species. For example, the human genome sequence in principle could be determined from just half the information on the DNA of one cell from one individual. To learn what variations in genetic information underlie particular traits or diseases requires comparisons across individuals. This point explains the common usage of "genome" (which parallels a common usage of "gene") to refer not to the information in any particular DNA sequence, but to a whole family of sequences that share a biological context. Although this concept may seem counter intuitive, it is the same concept that says there is no particular shape that is the shape of a cheetah. Cheetahs vary, and so do the sequences of their genomes. Yet both the individual animals and their sequences share commonalities, so one can learn something about cheetahs and "cheetah-ness" from a single example of either. 2.3 Characteristic of genome structure 2.3.1 Virus A virus (from the Latin virus meaning "toxin" or "poison"), is a sub-microscopic infectious agent that is unable to grow or reproduce outside a host cell. Each viral particle, or virion, consists of genetic material, DNA or RNA, within a protective protein coat called a capsid. The capsid shape varies from simple helical and icosahedral (polyhedral or near-spherical) forms, to more complex structures with tails or an envelope. Viruses infect cellular life forms and are grouped into animal, plant and bacterial types, according to the type of host infected. An enormous variety of genomic structures can be seen among viral species; as a group they contain more structural genomic diversity than the entire kingdoms of either plants, animals, or bacteria. 2.3.1.1 Size. Genome size in terms of the weight of nucleotides varies between species. The smallest genomes code for only four proteins and weigh about 106 Daltons, the largest weigh about 108 Daltons and code for over one hundred proteins. RNA viruses generally have smaller genome sizes than DNA viruses due to a higher error-rate when replicating, resulting in a maximum upper size limit. Beyond this limit, errors in the genome when replicating render the virus useless or uncompetitive. To compensate for this, RNA viruses often have segmented genomes where the genome is split into smaller molecules, thus reducing the chance of error. In contrast, DNA viruses generally have larger genomes due to the high fidelity of their replication enzymes. 2.3.1.2 Genome. A virus may employ either DNA or RNA as the nucleic acid. Rarely do they contain both, however cytomegalovirus is an exception to this, possessing a DNA core with several mRNA segments. By far most viruses have RNA. Plant viruses tend to have single-stranded RNA and bacteriophages tend to have double-stranded DNA. Some virus species possess abnormal nucleotides, such as hydroxymethylcytosine instead of cytosine, as a normal part of their genome. Viral genomes may be circular, such as polyomaviruses, or linear, such as adenoviruses. The type of nucleic acid is irrelevant to the shape of the genome

Among RNA viruses,the genome is often divided up into separate parts within the virion and are called segmented.Double-stranded RNA genomes and some single-stranded RNA ge omes ae segmented.Each on protein and they are usually found together in one capsid.Every segment is not required to be in the same virion for the overall virus to be infectious,as demonstrated by the brome mosaic virus. A viral genome,irrespective of nucleic acid type,may be either single-stranded or double-s strandeds nded g ome st of ar unpaired nu leic acid. analogous to one-half of a ladder split down the middle.Double-stranded genomes consist of 2 complementary paired nucleic acids,analogous to a ladder.Viruses. such as those belonging to the Hepadnaviridae,contain a genome which is partially double-stranded and partially single-stranded.Viruses that infect humans include double-stranded RNA(e.g.Rota virus),single-stranded RNA(e.g.Influenza virus), single-stranded DNA(e.g.Parvovirus B19)and double-stranded DNA(Herpes virus). For viruses with rNa as their nucleic acid.the strands are said to be either positive-sense(called the plus-strand)or negative-sense(called the minus-strand). depending on whether it is complementary to viral mRNA.Positive-sense viral RNA RNA and thus can be immediat ely tr ranslated by the host cell RA complncnay to mRNAnd to positive-sense RNA by an RNA polymerase before translation.DNA nomenclature is similar to RNA nomenclature,in that the coding strand for the viral mRNA is complementary to it(-),and the non-coding strand is a copy of it(+). 2.3.2 Prokaryotic 96 ogists recognize tha the living world comprises two types of organism 1.Eukaryotes,whose cells contain membrane-bound compartments,including a nucleus and organelles such as mitochondria and.in the case of plant cells. chloroplasts.Eukaryotes include animals,plants,fungi and protozoa. 2.Prokaryotes,whose cells lack extensive internal compartments.There are two groups of proka aryotes,distinguished from one another by characteristic a.the bacteria.which include most of the commonly encountered prokarvotes such as the gram-negatives(e.g.E.coli),the gram-positives(e.g.Bacillus subtilis),the cvanobacteria (e g anabaena)and many more: b.the arc which are less well-studied,and have mostly been found in extreme environments such as hot springs,brine pools and anaerobic lake bottoms 2.3.2.1 The size of the Prokaryotic genome.Prokaryotes have smaller,simpler genomes than eukaryotes.Most prokaryotic genomes are less than 5 Mb in size, although a few are substantially larger than this:B.megaterium,for example,has a huge n is a regior of cytoplasm ofp prokar e less dense tha surrounding cytoplasm.Prokaryotes may also have smaller,extrachromosomal DNA plasmids,that consist of only a few to a few hundreds of genes.Plasmids are non-essential (by definition):chromosomes are essential.However.plasmids often
12 Among RNA viruses, the genome is often divided up into separate parts within the virion and are called segmented. Double-stranded RNA genomes and some single-stranded RNA genomes are segmented. Each segment often codes for one protein and they are usually found together in one capsid. Every segment is not required to be in the same virion for the overall virus to be infectious, as demonstrated by the brome mosaic virus. A viral genome, irrespective of nucleic acid type, may be either single-stranded or double-stranded. Single-stranded genomes consist of an unpaired nucleic acid, analogous to one-half of a ladder split down the middle. Double-stranded genomes consist of 2 complementary paired nucleic acids, analogous to a ladder. Viruses, such as those belonging to the Hepadnaviridae, contain a genome which is partially double-stranded and partially single-stranded. Viruses that infect humans include double-stranded RNA (e.g. Rotavirus), single-stranded RNA (e.g. Influenza virus), single-stranded DNA (e.g. Parvovirus B19) and double-stranded DNA (Herpes virus). For viruses with RNA as their nucleic acid, the strands are said to be either positive-sense (called the plus-strand) or negative-sense (called the minus-strand), depending on whether it is complementary to viral mRNA. Positive-sense viral RNA is identical to viral mRNA and thus can be immediately translated by the host cell. Negative-sense viral RNA is complementary to mRNA and thus must be converted to positive-sense RNA by an RNA polymerase before translation. DNA nomenclature is similar to RNA nomenclature, in that the coding strand for the viral mRNA is complementary to it (-), and the non-coding strand is a copy of it (+). 2.3.2 Prokaryotic genomes Biologists recognize that the living world comprises two types of organism: 1. Eukaryotes, whose cells contain membrane-bound compartments, including a nucleus and organelles such as mitochondria and, in the case of plant cells, chloroplasts. Eukaryotes include animals, plants, fungi and protozoa. 2. Prokaryotes, whose cells lack extensive internal compartments. There are two very different groups of prokaryotes, distinguished from one another by characteristic genetic and biochemical features: a. the bacteria, which include most of the commonly encountered prokaryotes such as the gram-negatives (e.g. E. coli), the gram-positives (e.g. Bacillus subtilis), the cyanobacteria (e.g. Anabaena) and many more; b. the archaea, which are less well-studied, and have mostly been found in extreme environments such as hot springs, brine pools and anaerobic lake bottoms. 2.3.2.1 The size of the Prokaryotic genome. Prokaryotes have smaller, simpler genomes than eukaryotes. Most prokaryotic genomes are less than 5 Mb in size, although a few are substantially larger than this: B. megaterium, for example, has a huge genome of 30 Mb. Nucleoid region is a region of cytoplasm of prokaryotes which contains concentrated snarl of DNA fibers, and stains less dense than surrounding cytoplasm. Prokaryotes may also have smaller, extrachromosomal DNA, plasmids, that consist of only a few to a few hundreds of genes. Plasmids are non-essential (by definition); chromosomes are essential. However, plasmids often

provide the hosts certain advantages:e.g.,resistance to antibiotics,metabolism of nieids replicaie independenby ween partners during conjugation. 2.3.2.1 The genetic organization of the prokaryotic genome.We have already learnt that bacterial genomes have compact genetic organizations with very little space between genes(see Figure 2.1).There is non-coding DNA in the E.coli genome,but it accounts for only 11%of the total and it is distributed around the ome in sr segments that do not show up when the map is drawn at this scale In this regard,E.coli is typical of all prokaryotes whose genomes have so far been sequenced -prokaryotic genomes have very little wasted space.There are theories that this compact organization is beneficial to prokaryotes,for example by enabling the genome to be replicated relatively quickly,but these ideas have never been supported by hard experimental evidenc A 8)5 mT1mnp省i” 2 (C)Dres 早甲n,I可 nnmn首mm 即a Sc。on n口Cenome-wide repeat tANA gne Figure 2.1.Comparison of the genomes of humans,yeast,fruit flies,maize and Escherichia coli.(A)is the 50-kb segment of the human B T-cell receptor locus This is compared with 50-kb segments from the genomes of(B) revisiae(chro some III: edrawn from):(C)Drosophil melanogaster;(D)maize and (E)E.coli K12 One characteristic feature of prokaryotic genomes illustrated by E.coli is the presence of operons.An operon is a group of genes that are located adjacent to one another in the genome st one or two nucleotides be ween the end of one gene and the start of the next.All the genes in an operon are expressed as a single unit.This type of arrangement is common in prokaryotic genomes.A typical E.coli example is the lactose operon,the first operon to be discovered,which contains three genes involved in conversion of the disaccharide sugar lactose into its
13 provide the hosts certain advantages: e.g., resistance to antibiotics, metabolism of unusual nutrients, and other special contingencies. Plasmids replicate independently of the chromosome and can be transferred between partners during conjugation. 2.3.2.1 The genetic organization of the prokaryotic genome. We have already learnt that bacterial genomes have compact genetic organizations with very little space between genes (see Figure 2.1 ). . There is non-coding DNA in the E. coli genome, but it accounts for only 11% of the total and it is distributed around the genome in small segments that do not show up when the map is drawn at this scale. In this regard, E. coli is typical of all prokaryotes whose genomes have so far been sequenced - prokaryotic genomes have very little wasted space. There are theories that this compact organization is beneficial to prokaryotes, for example by enabling the genome to be replicated relatively quickly, but these ideas have never been supported by hard experimental evidence. Figure 2.1. Comparison of the genomes of humans, yeast, fruit flies, maize and Escherichia coli. (A) is the 50-kb segment of the human β T-cell receptor locus . This is compared with 50-kb segments from the genomes of (B) Saccharomycescerevisiae (chromosome III; redrawn from); (C) Drosophila melanogaster ; (D) maize and (E) E. coli K12 One characteristic feature of prokaryotic genomes illustrated by E. coli is the presence of operons. An operon is a group of genes that are located adjacent to one another in the genome, with perhaps just one or two nucleotides between the end of one gene and the start of the next. All the genes in an operon are expressed as a single unit. This type of arrangement is common in prokaryotic genomes. A typical E. coli example is the lactose operon, the first operon to be discovered, which contains three genes involved in conversion of the disaccharide sugar lactose into its

monosaccharide units-glucose and galactose(Figure 2.2 A).The monosaccharides are substrates for the energy-generating glycolytic pathway,so the function of the nes in the lactose operon is to convert lactose into a form tha an be utilized by coli as an energy source.Lactose is not a common component of E.colsnatural environment,so most of the time the operon is not expressed and the enzymes for lactose utilization are not made by the bacterium.When lactose becomes available,it switches on the operon;all three genes are expressed together,resulting in coordinated synthesis of the lacto utilizingen each containing two or more genes,and a similar number are present in Bacillus subtilis. In most cases the genes in an operon are functionally related,coding for a set of ed i single biohmicl acivy h synthesis of an amino acid.An example of the latter is the tryptophan operon ofE.co(Figure 22B).Microbial geneticists are attracted to the simplicity of this system whereby a bacterium is able to control its various biochemical activities by regulating the expression of groups of related genes linked together in operons.This may be a correct interpretation of the function of operons in many other prokaryotes bu in at least ome species the picture is ess.Both the areha the bacterium Aquifex aeolicus have operons,but the genes in an individual operon rarely have any biochemical relationship.For example,one of the operons in the A. aeolicus genome contains six linked genes,these genes coding for two proteins involved in DNA recombination,an enzyme used in protein synthesis,a protein uired for motility. esis and an 3ure2.37 involved in nucleotide synth This is typical of the operon structure in the and M.jannaschii genomes.In other words,the notion that expression of an operon leads to the coordinated synthesis of enzymes required for a single biochemical pathway does not hold for these species
14 monosaccharide units - glucose and galactose ( Figure 2.2 A ). The monosaccharides are substrates for the energy-generating glycolytic pathway, so the function of the genes in the lactose operon is to convert lactose into a form that can be utilized by E. coli as an energy source. Lactose is not a common component of E. coli's natural environment, so most of the time the operon is not expressed and the enzymes for lactose utilization are not made by the bacterium. When lactose becomes available, it switches on the operon; all three genes are expressed together, resulting in coordinated synthesis of the lactose-utilizing enzymes. Altogether there are almost 600 operons in the E. coli K12 genome, each containing two or more genes, and a similar number are present in Bacillus subtilis. In most cases the genes in an operon are functionally related, coding for a set of proteins that are involved in a single biochemical activity such as utilization of a sugar as an energy source or synthesis of an amino acid. An example of the latter is the tryptophan operon of E. coli ( Figure 2.2B ). Microbial geneticists are attracted to the simplicity of this system whereby a bacterium is able to control its various biochemical activities by regulating the expression of groups of related genes linked together in operons. This may be a correct interpretation of the function of operons in E. coli, Bacillus subtilis and many other prokaryotes, but in at least some species the picture is less straightforward. Both the archaeon Methanococcus jannaschii and the bacterium Aquifex aeolicus have operons, but the genes in an individual operon rarely have any biochemical relationship. For example, one of the operons in the A. aeolicus genome contains six linked genes, these genes coding for two proteins involved in DNA recombination, an enzyme used in protein synthesis, a protein required for motility, an enzyme involved in nucleotide synthesis, and an enzyme for lipid synthesis ( Figure 2.3 ). This is typical of the operon structure in the A. aeolicus and M. jannaschii genomes. In other words, the notion that expression of an operon leads to the coordinated synthesis of enzymes required for a single biochemical pathway does not hold for these species
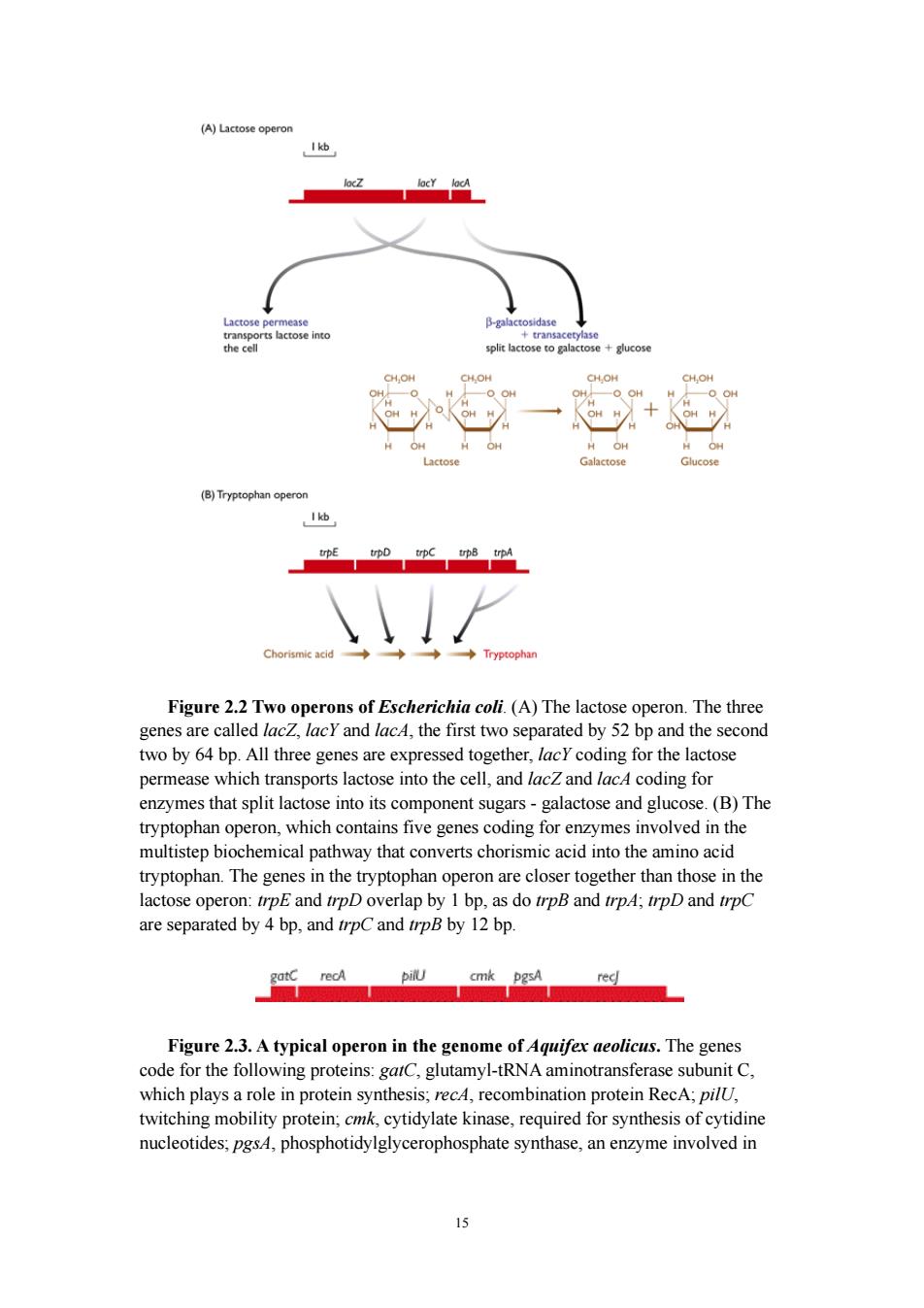
cohatnmo @一2 Chorismic acid◆ Figure 2.2Two operons of Escherichia coli (A)The lactose operon.The three genes are called lacZ,lacY and lacA,the first two separated by 52 bp and the second two by 64 bp.All three genes are expressed together,lacY coding for the lactose permease which transports lactose into the cell,and lacZ and lac4 coding for enzymes that split lactose into its component sug -galactose and glu ose.(B)The tryptophan operon,which contains five genes coding for enzymes involved in the multistep biochemical pathway that converts chorismic acid into the amino acid tryptophan.The genes in the tryptophan operon are closer together than those in the lactose operon:trpE and trpD overlap by 1 bp,as do trpB and trpA:trpD and trpC are separated by 4 bp,and trpC and trpB by 12 bp. cmk pgs Figure 2.3.A typical operon in the genome of Aquifex aeolicus.The genes wing proteins:gatC,glutamyl-tRNAa ubunit C. oe in protein synthesis:rec,recomtion protein RecA twitching mobility protein;mk cytidylate kinase,required for synthesis of cytidine nucleotides,pgsA,phosphotidylglycerophosphate synthase,an enzyme involved in
15 Figure 2.2 Two operons of Escherichia coli. (A) The lactose operon. The three genes are called lacZ, lacY and lacA, the first two separated by 52 bp and the second two by 64 bp. All three genes are expressed together, lacY coding for the lactose permease which transports lactose into the cell, and lacZ and lacA coding for enzymes that split lactose into its component sugars - galactose and glucose. (B) The tryptophan operon, which contains five genes coding for enzymes involved in the multistep biochemical pathway that converts chorismic acid into the amino acid tryptophan. The genes in the tryptophan operon are closer together than those in the lactose operon: trpE and trpD overlap by 1 bp, as do trpB and trpA; trpD and trpC are separated by 4 bp, and trpC and trpB by 12 bp. Figure 2.3. A typical operon in the genome of Aquifex aeolicus. The genes code for the following proteins: gatC, glutamyl-tRNA aminotransferase subunit C, which plays a role in protein synthesis; recA, recombination protein RecA; pilU, twitching mobility protein; cmk, cytidylate kinase, required for synthesis of cytidine nucleotides; pgsA, phosphotidylglycerophosphate synthase, an enzyme involved in