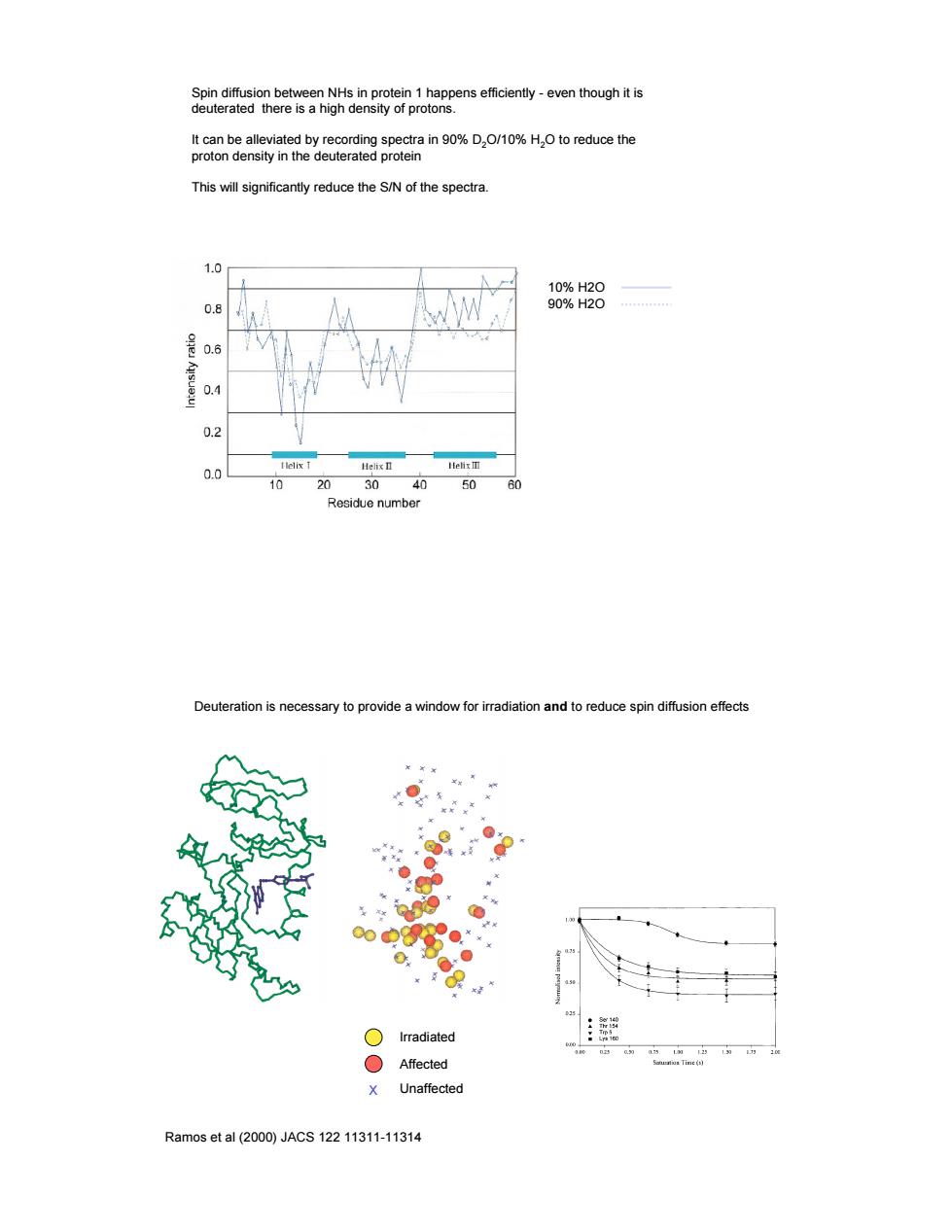
SenemeaombesgnhienmS28rploieRpense6ceny-eventhoughts t can be alleviated by recording spectra in 90%D.O/10%H.O to reduce the proton density in the deuterated protein This will significantly reduce the S/N of the spectra. n a 98%8 0.6 0.4 03 0 10 20 30 50 60 Residue number erauon is ne adiation and to reduce spin diffusion effects ○Irradiated Affected X Unaffected Ramos et al(2000)JACS12211311-11314
Spin diffusion between NHs in protein 1 happens efficiently - even though it is deuterated there is a high density of protons. It can be alleviated by recording spectra in 90% D2O/10% H2O to reduce the proton density in the deuterated protein This will significantly reduce the S/N of the spectra. 10% H2O 90% H2O Deuteration is necessary to provide a window for irradiation and to reduce spin diffusion effects Ramos et al (2000) JACS 122 11311-11314 X Irradiated Affected Unaffected

nces in the H spectrum at-6 ppm (H1'H5)and 12 ppm (indole groups) 载 Crystal structure RNA contacts Cross-saturation Chemical shift perturbation Ramos et al(2000)JACS12211311-11314 Distance Information
Ramos et al (2000) JACS 122 11311-11314 Nucleic acids have resonances in the 1H spectrum at ~6 ppm (H1’,H5) and 12 ppm (indole groups) Many proteins do not. Crystal structure RNA contacts Cross-saturation Chemical shift perturbation Distance Information
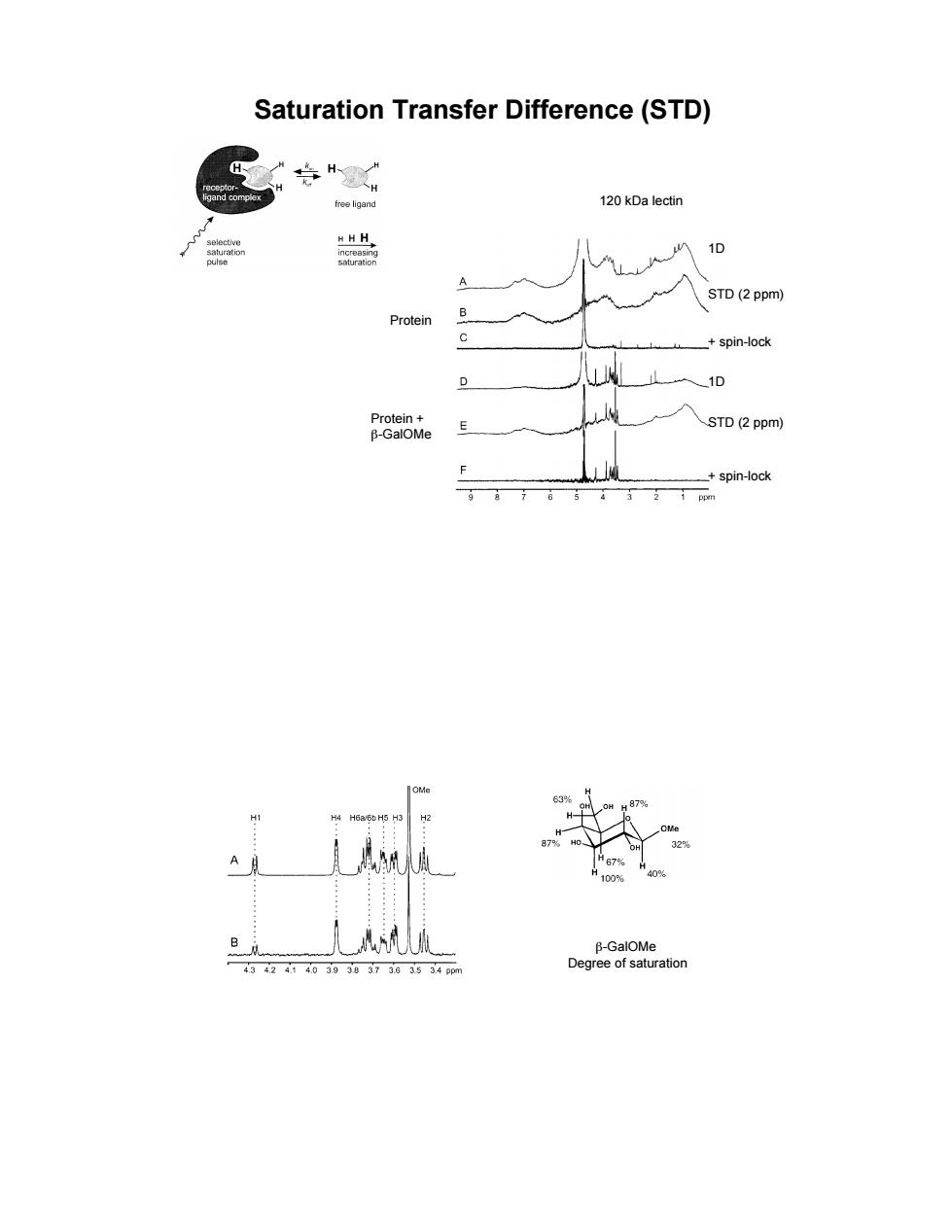
Saturation Transfer Difference(STD) 120 kDa lectin A STD(2ppm) Protein B _+spin-ock 0 STD (2 ppm) spin-ock
Saturation Transfer Difference (STD) 120 kDa lectin 1D STD (2 ppm) + spin-lock 1D STD (2 ppm) + spin-lock Protein Protein + &-GalOMe &-GalOMe Degree of saturation
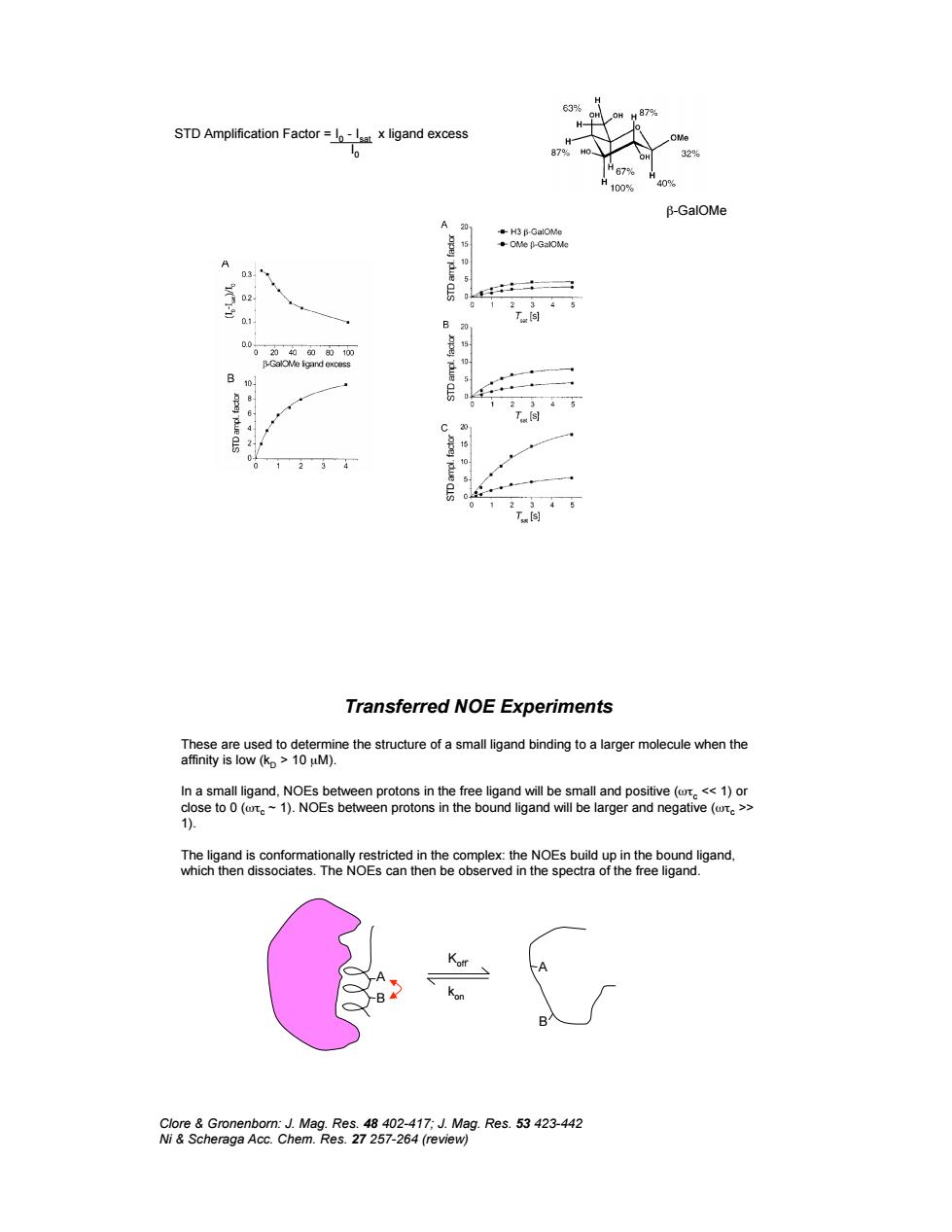
STD Amplification Factorx ligand excess Transferred NOE Experiments o96 ine the structure of a small ligand binding to a larger molecule when the ÷
&-GalOMe STD Amplification Factor = I 0 - Isat x ligand excess I 0 Transferred NOE Experiments These are used to determine the structure of a small ligand binding to a larger molecule when the affinity is low (kD > 10 µM). In a small ligand, NOEs between protons in the free ligand will be small and positive ('%c << 1) or close to 0 ('%c ~ 1). NOEs between protons in the bound ligand will be larger and negative ('%c >> 1). The ligand is conformationally restricted in the complex: the NOEs build up in the bound ligand, which then dissociates. The NOEs can then be observed in the spectra of the free ligand. Clore & Gronenborn: J. Mag. Res. 48 402-417; J. Mag. Res. 53 423-442 Ni & Scheraga Acc. Chem. Res. 27 257-264 (review) A B A B Koff` kon

Transferred NOE Experiments K A A-B tr NOE NOE r NOE A-B NOE relayed via spin diffusion Direct NOE (A.-B) Mixing time )ms Ni&Scheraga Acc.Chem.Res.27257-264 Transferred NOEs 29cme6tgandmawacaedsocaeaevnemg -NOE build-up curves are used to optimize the mixing time Intermolecular NOEs may also be observed on rates(aso time) -53
Transferred NOE Experiments Ni & Scheraga Acc. Chem. Res. 27 257-264 Ab Bb Af Bf Koff` kon Z Mixing time NOE 600 ms A-B tr NOE Intermolecular tr NOE (Z-B) A-B NOE relayed via spin diffusion Direct NOE (Ab-Bb) 100% 0% 4 types of NOE are possible Transferred NOEs •Ligand must be in large excess over protein - at least 10X, so the protein is saturated with ligand but the linewidths are dominated by free ligand •The off-rate must be fast: the ligand must associate/dissociate a few times during the NOE mixing time •NOE build-up curves are used to optimize the mixing time •Intermolecular NOEs may also be observed •Transferred cross correlated relaxation rates (also depend on correlation time) (Blommers et al 1999 JACS 121 1949-53 Carlomagno 1999 JACS 121 1945-48) •Transferred dipolar couplings have also been observed (special case if protein is weakly aligned) (Koenig et al 2000 J.Biomol. NMR 16 121-125)