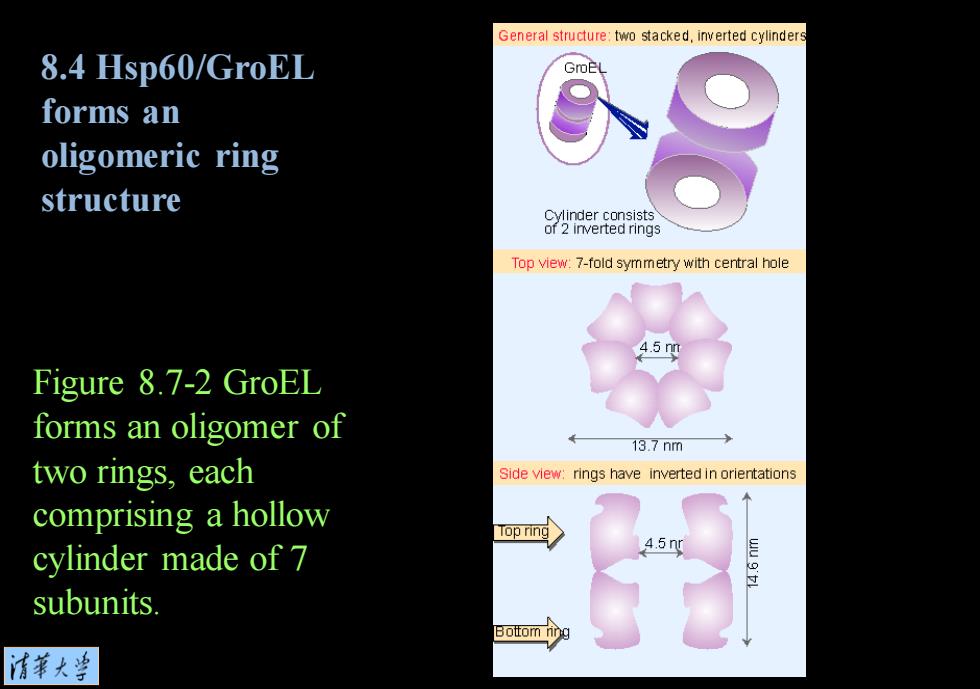
General structure:two stacked,inverted cylinders 8.4 Hsp60/GroEL forms an oligomeric ring structure 2Wmverteumng Top view:7-fold symmetry with central hole Figure 8.7-2 GroEL forms an oligomer of 13.7nm two rings,each Side view:rings have inverted in orientations comprising a hollow Top ring cylinder made of 7 4.5ny subunits 清菜大当
Figure 8.7-2 GroEL forms an oligomer of two rings, each comprising a hollow cylinder made of 7 subunits. 8.4 Hsp60/GroEL forms an oligomeric ring structure
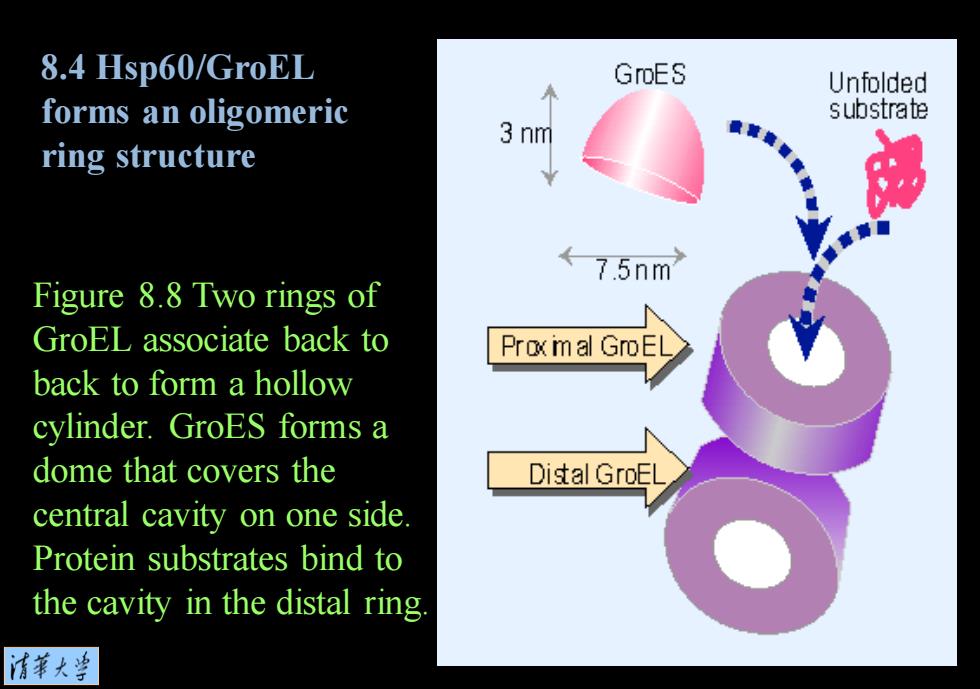
8.4 Hsp60/GroEL GroES Unfolded forms an oligomeric substrate 3 nm ring structure ←75nm Figure 8.8 Two rings of GroEL associate back to Proxm al GroEL back to form a hollow cylinder.GroES forms a dome that covers the Distal GroEL central cavity on one side. Protein substrates bind to the cavity in the distal ring 清菜大当
Figure 8.8 Two rings of GroEL associate back to back to form a hollow cylinder. GroES forms a dome that covers the central cavity on one side. Protein substrates bind to the cavity in the distal ring. 8.4 Hsp60/GroEL forms an oligomeric ring structure
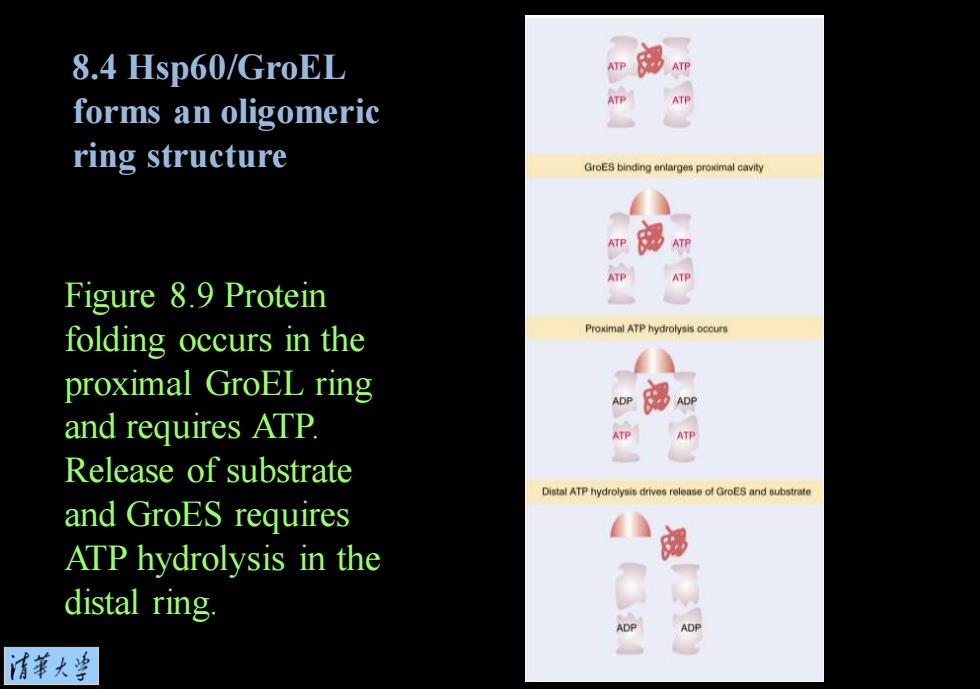
8.4 Hsp60/GroEL forms an oligomeric ATP ATP ring structure GroES binding enlarges proxmal cavity ATP 部 ATP Figure 8.9 Protein folding occurs in the Proximal ATP hydrolysis occurs proximal GroEL ring o部e and requires ATP. ATP ATP Release of substrate and GroES requires ATP hydrolysis in the distal ring. ADP ADP 情菜大兰
Figure 8.9 Protein folding occurs in the proximal GroEL ring and requires ATP. Release of substrate and GroES requires ATP hydrolysis in the distal ring. 8.4 Hsp60/GroEL forms an oligomeric ring structure

ORGANELLE 8.5 Post-translational Pmtein passes through 的已nren惊a3过ěa0eG membrane insertion 5白9uf0e5ee3We0 depends on leader ature protein sequences Figure 8.10 Leader sequences allow proteins to recognize Leader sequence binds mitochondrial or to receptor on organelle chloroplast surfaces membrane by a post-translational CYTOSOL process. 清菜大当
Figure 8.10 Leader sequences allow proteins to recognize mitochondrial or chloroplast surfaces by a post-translational process. 8.5 Post-translational membrane insertion depends on leader sequences
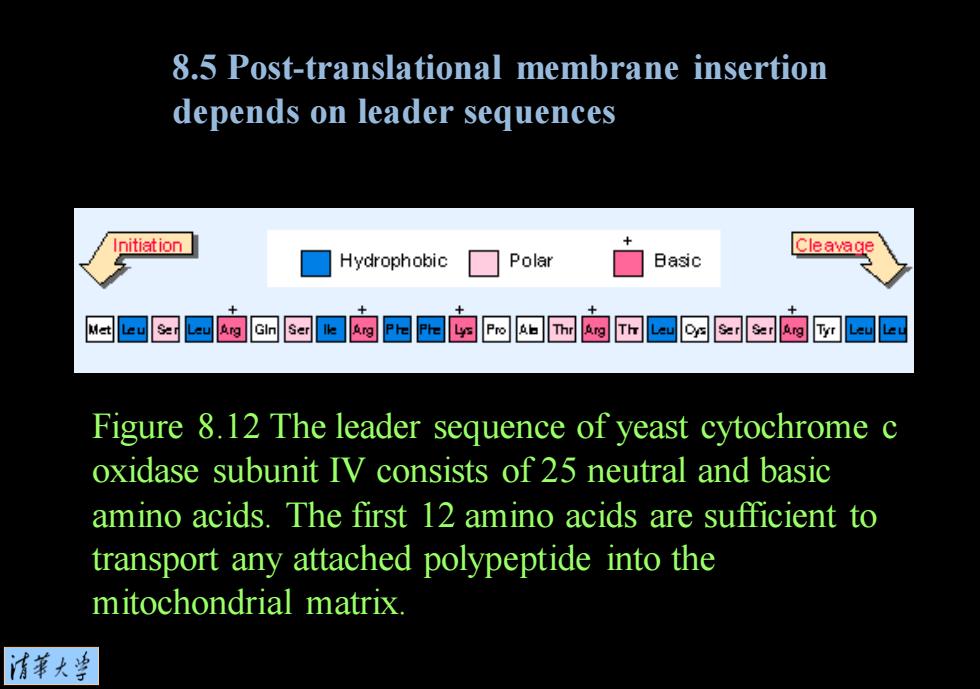
8.5 Post-translational membrane insertion depends on leader sequences Initiation Hydrophobic Polar Basic Cleavage 回匀回购厨▣购四四的四如园内而回函的团回回 Figure 8.12 The leader sequence of yeast cytochrome c oxidase subunit IV consists of 25 neutral and basic amino acids.The first 12 amino acids are sufficient to transport any attached polypeptide into the mitochondrial matrix. 情菜大兰
Figure 8.12 The leader sequence of yeast cytochrome c oxidase subunit IV consists of 25 neutral and basic amino acids. The first 12 amino acids are sufficient to transport any attached polypeptide into the mitochondrial matrix. 8.5 Post-translational membrane insertion depends on leader sequences