
EARLY AND MIDDLE 11.3 Functional DNA SYNTHESIS Replication clustering in 17 essential genes 7 nonessential genes LATE PHASE Modification ■ HEAD ASSEMBLY phages T7 and T4 3 nonessential genes Neck collar 1nonessenual gone DNA PRECURSORS Capsid components Host DNA breakdown 7 essential genes nonessential gene nonessential genes Figure 11.7 The phage T4 Capsid assembly Nucleotide metabolism 8tnTg8ne lytic cascade falls into two DNA packaging CELL STRUCTURE 昆82oteh88neg parts:early and quasi-late ■ Membrane functions ■ TAIL ASS日MBLY functions are concerned with 2 nonessential genes Baseplate components DNA synthesis and gene Lysis 13 essential genes ■ 2 nonessential genes ■ 日aseplate assembly expression;late functions are ■ 5 essential genes GENE EXPRESSION 2 nonessential genes concerned with particle Translation Tube she ath 1 2 nonessertial genes 4 essential gones assembly. Transcription Tail fibers 层8 essenuan88nes essential genes nonessential gene 清菜大兰
Figure 11.7 The phage T4 lytic cascade falls into two parts: early and quasi-late functions are concerned with DNA synthesis and gene expression; late functions are concerned with particle assembly. 11.3 Functional clustering in phages T7 and T4

bound by 11.3 Functional Cll alone clustering in bound by Cll +polymerase phages T7 and T4 出uu以 50-40-30-20-10 +10 Figure 11.24 RNA Startpoint polymerase binds Usual Usual to PRE only in the sequence sequence at-35 at-10 presence of CIl. TTGACA TATAAT which contacts the GCAACGCAAACAAACGT GCTT GGTATACATTCATAAAGGAATCTA CGTT GCGTTT GTTTGCACGAACCATAT GTAAGTATTTCCTTAGAT region around -35 ★大★大大 大★★大 cyR mutations cyL mutations polymerase binding polymerase binding 情菜大当 affect cll binding
Figure 11.24 RNA polymerase binds to PRE only in the presence of CII, which contacts the region around -35. 11.3 Functional clustering in phages T7 and T4
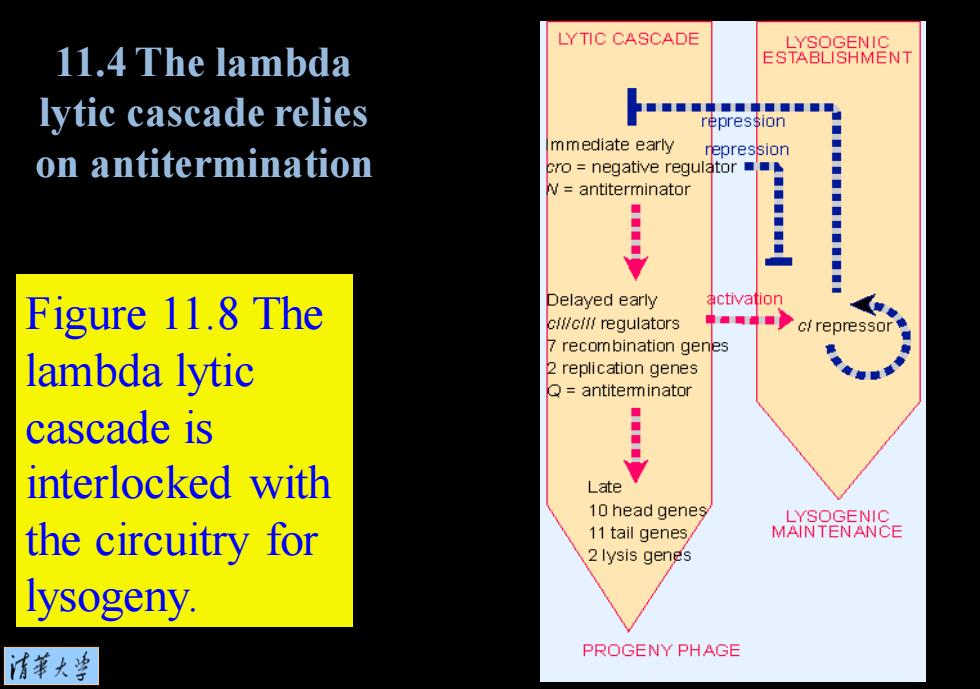
LYTIC CASCADE LYSOGENIC 11.4 The lambda ESTABLISHMENT lytic cascade relies ■■■■中■■中■■■■■ repression mmediate early repression ◆ on antitermination cro negative regulator : V antiterminator ◆ ◆ ◆ Figure 11.8 The Delayed early activation cli/cilf regulators c/repressor 7 recombination genes lambda lytic 2 replication genes Q=antitemminator ◆ cascade is interlocked with Late 10 head genes LYSOGENIC the circuitry for 11 tail genes, MAINTENANCE 2 lysis genes lysogeny. 清第大当 PROGENY PHAGE
Figure 11.8 The lambda lytic cascade is interlocked with the circuitry for lysogeny. 11.4 The lambda lytic cascade relies on antitermination
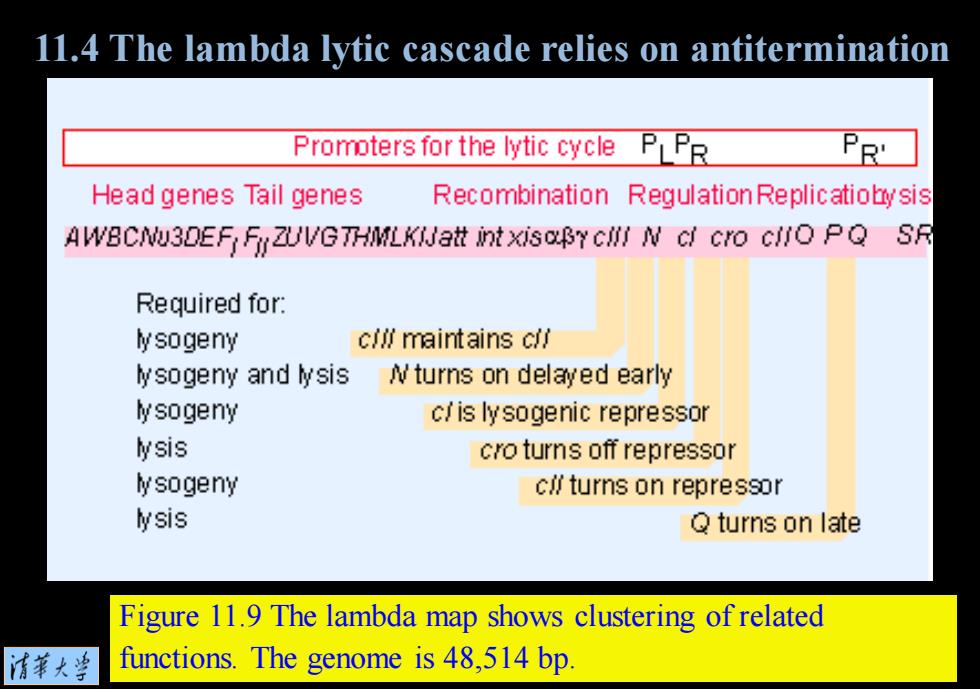
11.4 The lambda lytic cascade relies on antitermination Promoters for the lytic cycle PLPR Head genes Tail genes Recombination Regulation Replicatiobysis AWBCNU3DEF FyZUVGTHMLKIJatt int xisaBY CII N cl cro cilO PQ SR Required for: lysogeny cllf maintains clf lysogeny and lysis Nturns on delayed early lysogeny c/is lysogenic repressor lysis cro turns off repressor lysogeny c∥turns on repressor lysis Q turns on late Figure 11.9 The lambda map shows clustering of related 清菜大当 functions.The genome is 48,514 bp
Figure 11.9 The lambda map shows clustering of related functions. The genome is 48,514 bp. 11.4 The lambda lytic cascade relies on antitermination