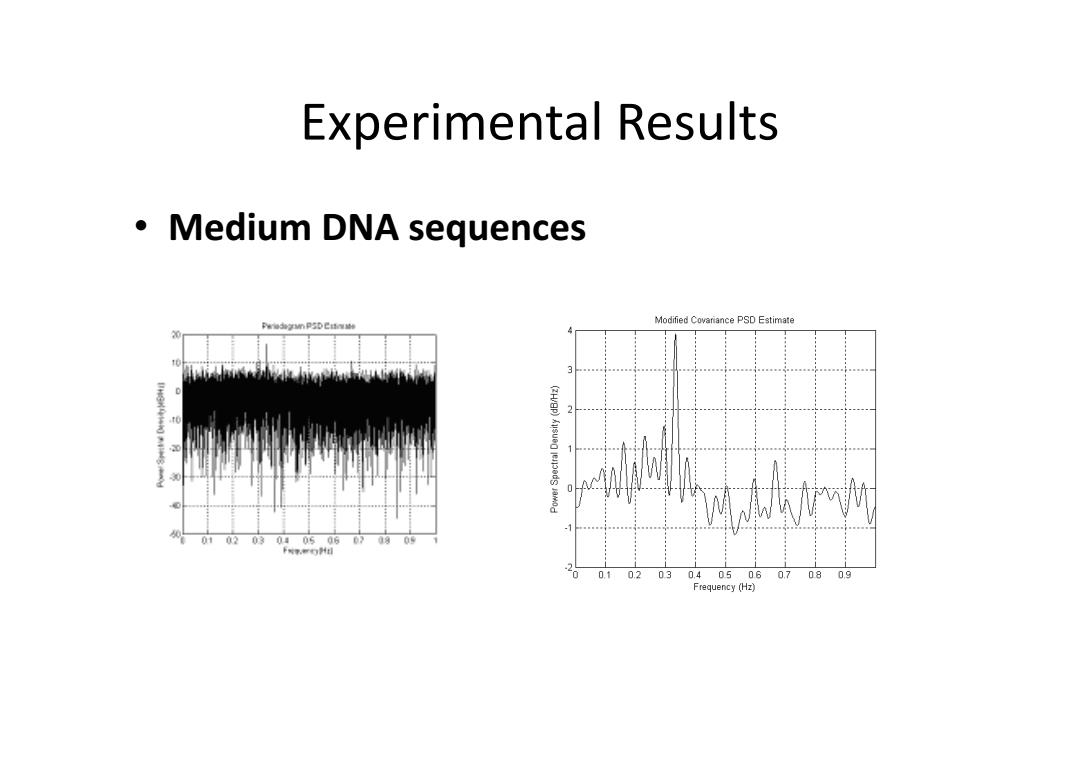
Experimental Results ·Medium DNA sequences Modified Covariance PSD Estimate Pu3海nPDC制 010203040506 09 卡对.门4目 20 0.1 02030.40506 07080g Frequency (Hz)
Experimental Results • Medium DNA sequences
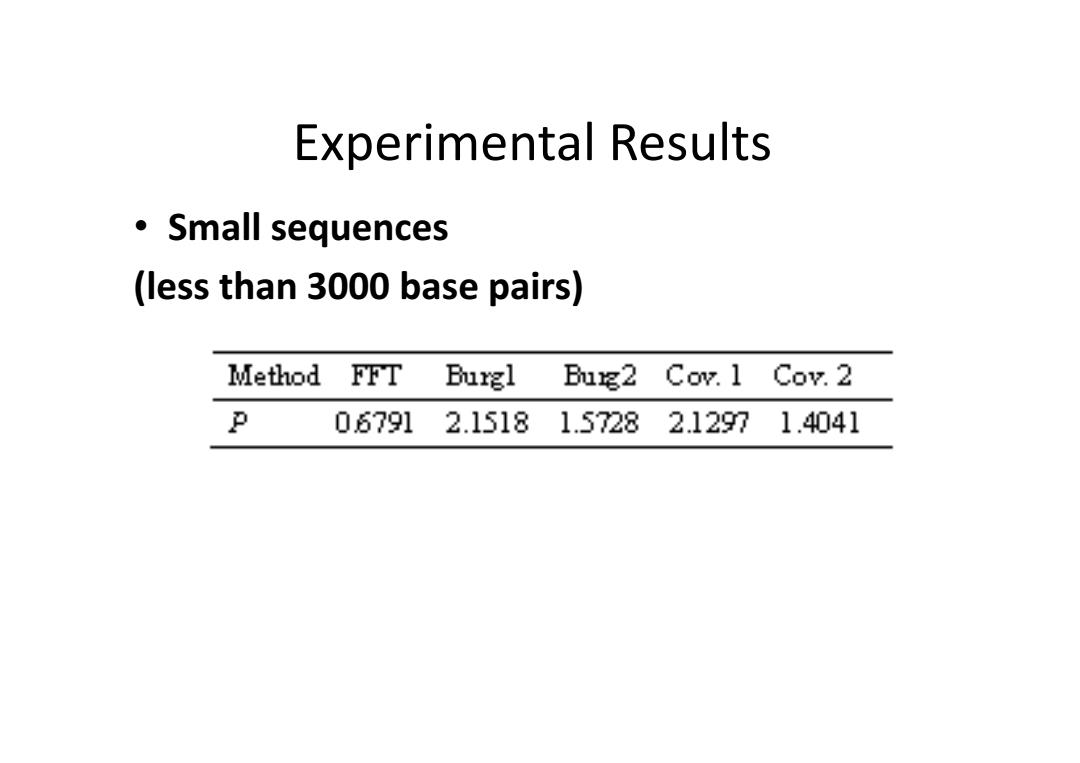
Experimental Results ·Small sequences (less than 3000 base pairs) Method FFT Burgl Bug2 Cov.1 Cov.2 P 06791 2.15181.57282.12971.4041
Experimental Results • Small sequences (less than 3000 base pairs)
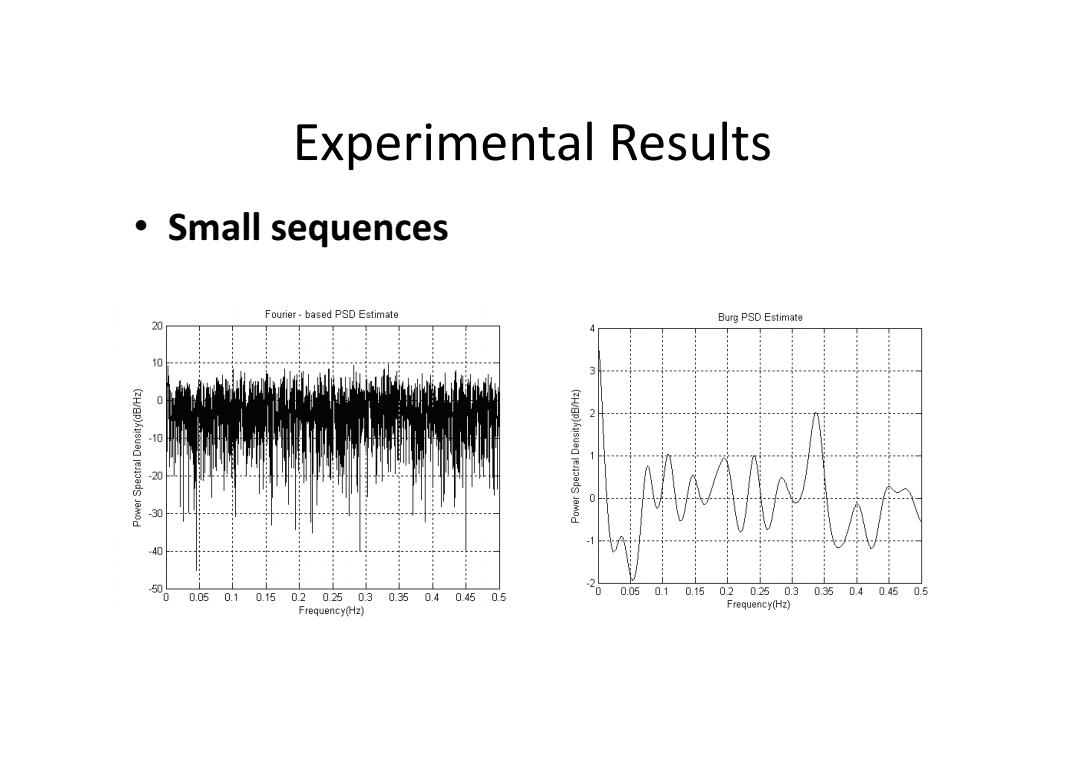
Experimental Results ·Small sequences Fourier-based PSD Estimate Burg PSD Estimate 0 i州n (ZH/EP)Aus 20 . 5 0 0.05 0.10.150.20250.30.350.40.450.5 0 0.050.10.150.20.250.30.350.40.450.5 Frequency(Hz) Frequency(Hz)
Experimental Results • Small sequences

Experimental Results The detection rates of five methods for short sequences Method FFT Burgl Burg2 Cov.1 Cov.2 Detection rate ( 1213%82.3% 73.1%82.6%75%
Experimental Results • The detection rates of five methods for short sequences
Discussion and conclusions AR methods are more robust The minimum detected length for satisfactory AR model performance is about 800 base pairs whereas it is around 2000 base pairs for the FFT method in the same case; Unlike neural net-based methods,no a prior knowledge of the nature or character of the sequences that constitute the coding region in a specific organism are required; The AR method is a useful tool for the analysis of sequences of unknown provenance
Discussion and conclusions • AR methods are more robust ; • The minimum detected length for satisfactory AR model performance is about 800 base pairs whereas it is around 2000 base pairs for the FFT method in the same case; • Unlike neural net–based methods, no a prior knowledge of the nature or character of the sequences that constitute the coding region in a specific organism are required; • The AR method is a useful tool for the analysis of sequences of unknown provenance