
9.4 Sigma factor controls Holoenzyme Equilibrium binding to DNA constant DNA binding Kg=105.109M1 Closed binary complex DNA melting Rate constant k2=103.101 sec Open Figure 9.9 RNA /A binary complex polymerase passes through Abortive several steps prior to initiation R69 sec elongation.A closed binary Comp complex is converted to an Release of Promoter open form and then into a sigma clearance >1-2 sec ternary complex RNA synthesis begins 清菜大当
Figure 9.9 RNA polymerase passes through several steps prior to elongation. A closed binary complex is converted to an open form and then into a ternary complex. 9.4 Sigma factor controls binding to DNA
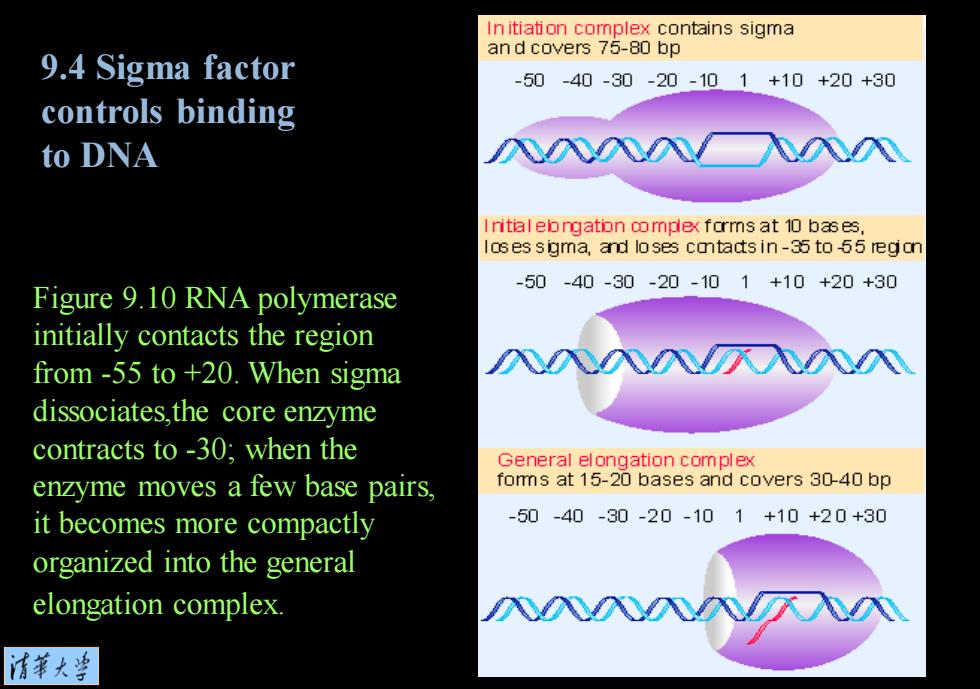
In itiation complex contains sigma and covers 75-80 bp 9.4 Sigma factor -50-40-30-20-101+10+20+30 controls binding to DNA Initial elongation oomplex forms at 10 bases, loses sigma,and loses catadts in-35 to -65 region Figure 9.10 RNA polymerase -50-40-30-20-101+10+20+30 initially contacts the region from -55 to +20.When sigma 入入入入八入入N dissociates,the core enzyme contracts to -30;when the General elongation complex enzyme moves a few base pairs, foms at 15-20 bases and covers 30-40 bp it becomes more compactly -50-40-30-20-101+10+20+30 organized into the general elongation complex. 清苇大当
Figure 9.10 RNA polymerase initially contacts the region from -55 to +20. When sigma dissociates,the core enzyme contracts to -30; when the enzyme moves a few base pairs, it becomes more compactly organized into the general elongation complex. 9.4 Sigma factor controls binding to DNA
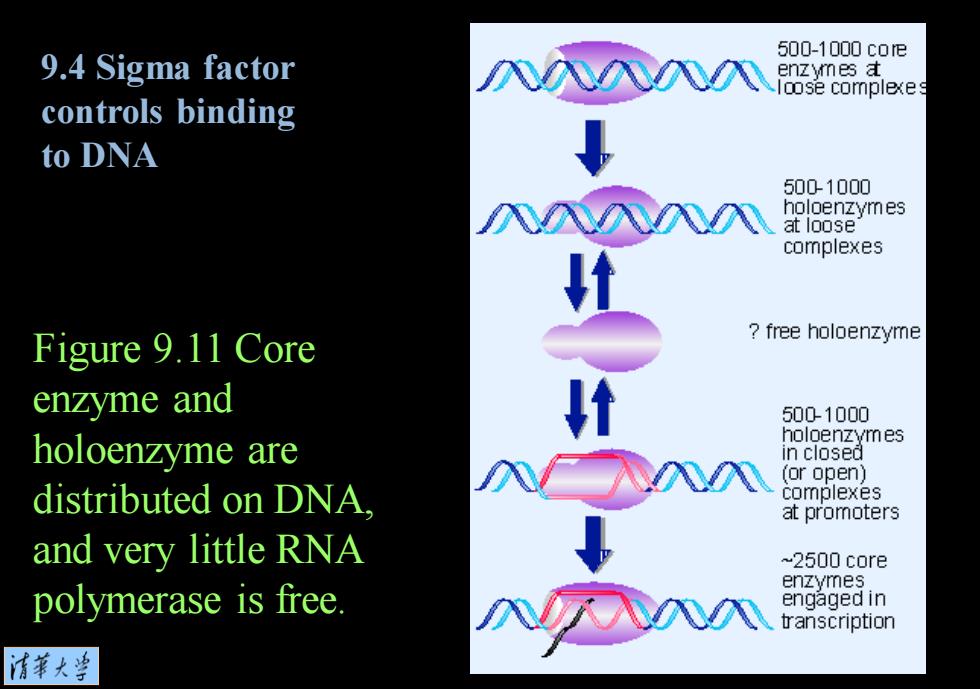
500-1000c0e 9.4 Sigma factor enzymes a loose comple以es controls binding to DNA 500-1000 holoenzymes at loose complexes Figure 9.11 Core free holoenzyme en☑yme and 5001000 holoenzyme are holoenzymes in closed (or open) distributed on DNA. complexes at promoters and very little RNA ~2500 core enzymes polymerase is free. engaged in transcription 情菜大兰
Figure 9.11 Core enzyme and holoenzyme are distributed on DNA, and very little RNA polymerase is free. 9.4 Sigma factor controls binding to DNA

Model Reaction 9.4 Sigma factor controls binding 八入入八N八 Random diffusion to DNA to target 109 Msec 八八八入N入 八八N入八N Random diffusion 戗64we to any DNA 八八N入八NN followed by 八入NNNN Random Figure 9.12 How does displace- ment between DNA N入闪入入入 RNA polymerase find N八八入八N target promoters so 八八入八入八 rapidly on DNA? Sliding along DNA Does not occur 清苇大当 N入入N入
Figure 9.12 How does RNA polymerase find target promoters so rapidly on DNA? 9.4 Sigma factor controls binding to DNA
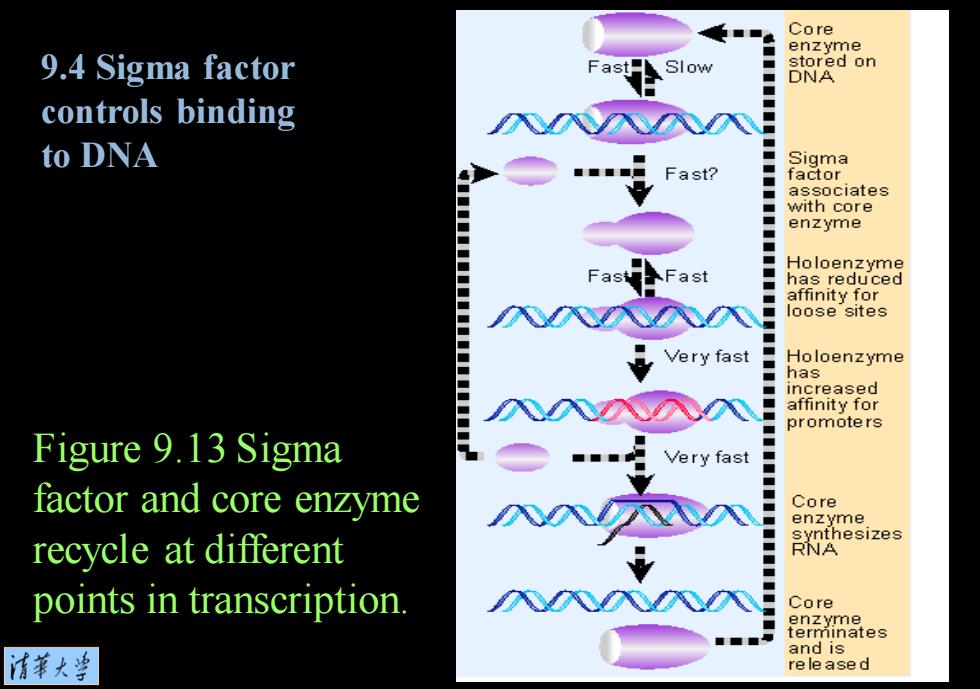
Core enzyme 9.4 Sigma factor Slow stored on DNA controls binding to DNA Sigma Fast? factor associates ■ with core ■ enzyme Holoenzyme -ast has reduced affinity for oose sites Very fast Holoenzyme has increased affinity for promoters Figure 9.13 Sigma factor and core enz☑yme % Very fast ■ Core enzyme recycle at different synthesizes RNA points in transcription. Core 情菜大当 and is released
Figure 9.13 Sigma factor and core enzyme recycle at different points in transcription. 9.4 Sigma factor controls binding to DNA