11.2 Sequencing and Annotating Genomes Sequencing:determining the precise order of nucleotides in a DNA or RNA molecule @Sanger dideoxy method桑格双脱氧测序方法 Invented by Nobel Prize winner Fred Sanger Dideoxy analogs of dNTPs used in conjunction with dNTPs Analog prevents further extension of DNA chain Bases are labeled with radioactivity Gel electrophoresis is then performed on products Chen Feng,Lecture of Microbiology
Chen Feng, Lecture of Microbiology 11.2 Sequencing and Annotating Genomes Sequencing: determining the precise order of nucleotides in a DNA or RNA molecule Sanger dideoxy method 桑格双脱氧测序方法 • Invented by Nobel Prize winner Fred Sanger • Dideoxy analogs of dNTPs used in conjunction with dNTPs • Analog prevents further extension of DNA chain • Bases are labeled with radioactivity • Gel electrophoresis is then performed on products
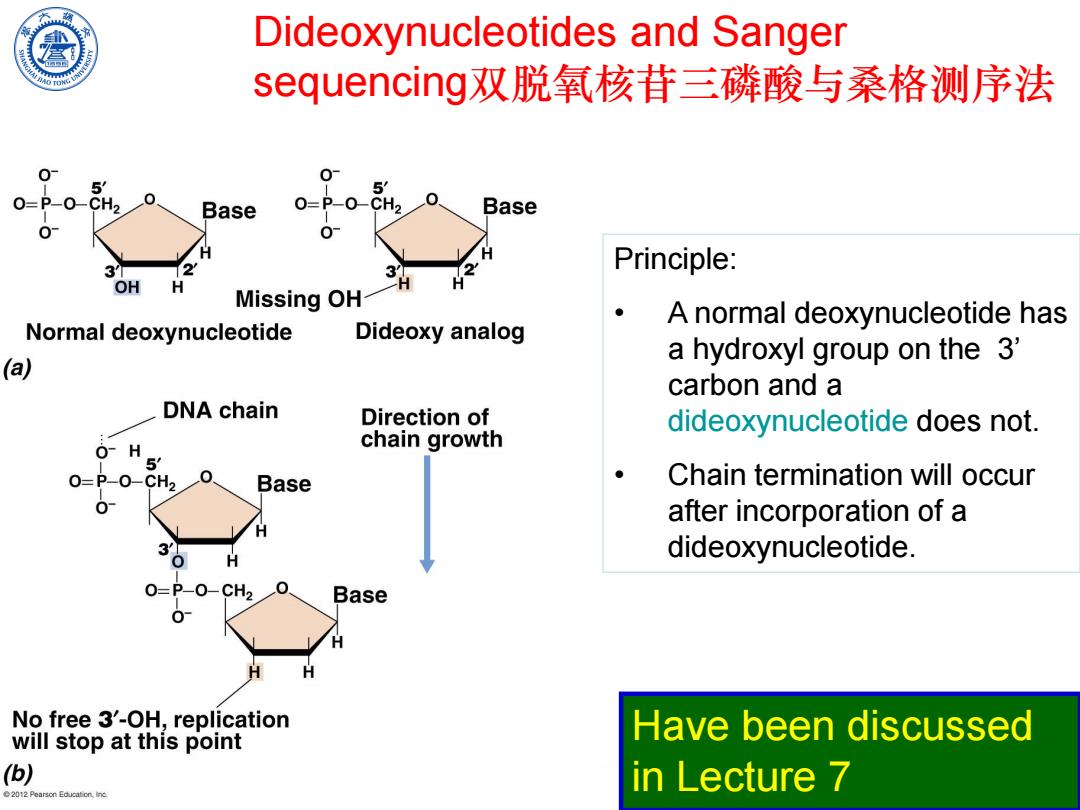
Dideoxynucleotides and Sanger sequencing双脱氧核苷三磷酸与桑格测序法 5 CH2 Base O=P-O-CH2 Base 0 3 Principle: OH H Missing OH- A normal deoxynucleotide has Normal deoxynucleotide Dideoxy analog (a) a hydroxyl group on the 3' carbon and a DNA chain Direction of dideoxynucleotide does not. O-H chain growth 5 0=P-0-CH20 Base Chain termination will occur 0 after incorporation of a dideoxynucleotide. P-O-CH2 Base No free 3'-OH,replication will stop at this point Have been discussed (b) in Lecture 7 2012 Pearson Educaton,ine
Chen Feng, Lecture of Microbiology Dideoxynucleotides and Sanger sequencing双脱氧核苷三磷酸与桑格测序法 Principle: • A normal deoxynucleotide has a hydroxyl group on the 3’ carbon and a dideoxynucleotide does not. • Chain termination will occur after incorporation of a dideoxynucleotide. Have been discussed in Lecture 7
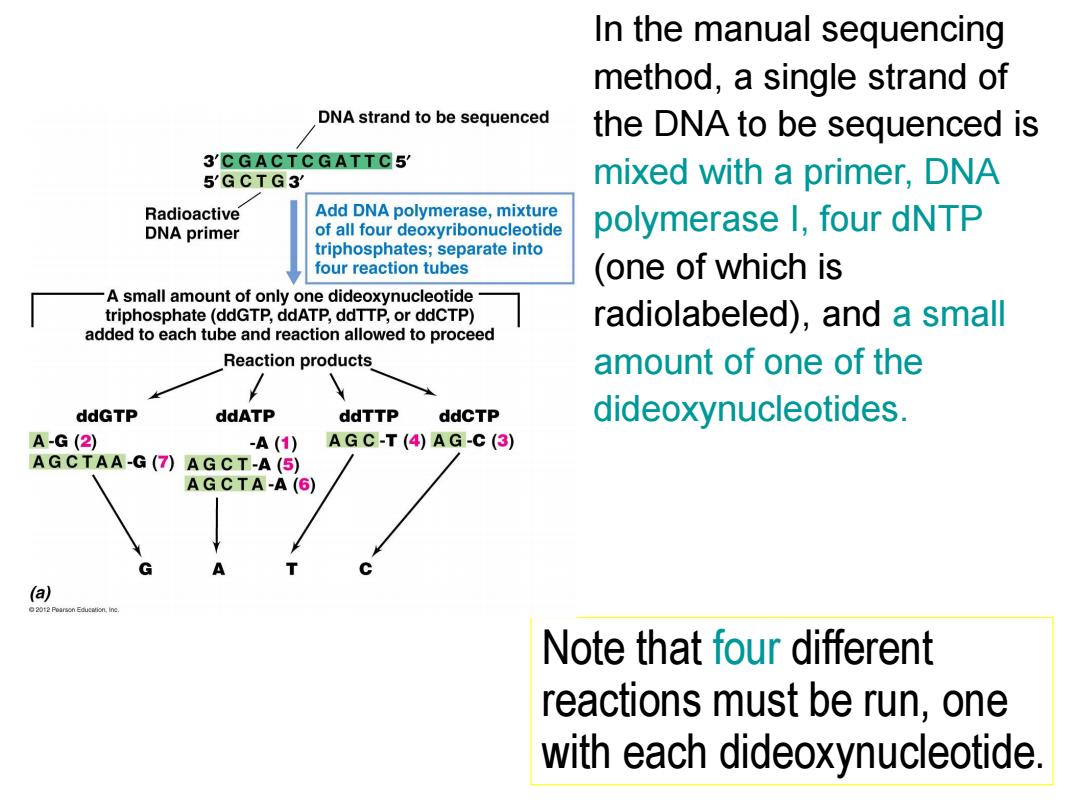
In the manual sequencing method,a single strand of DNA strand to be sequenced the DNA to be sequenced is 3'CGACTCGATTC5 5'GCTG3 mixed with a primer,DNA Radioactive Add DNA polymerase,mixture DNA primer of all four deoxyribonucleotide polymerase I,four dNTP triphosphates;separate into four reaction tubes (one of which is -A small amount of only one dideoxynucleotide triphosphate(ddGTP,ddATP,ddTTP,or ddCTP) radiolabeled),and a small added to each tube and reaction allowed to proceed Reaction products amount of one of the ddGTP ddATP ddTTP ddCTP dideoxynucleotides. A-G(2) -A(1) AGC-T(4)AG-C(3) AGCTAA-G(7)AGCT-A(5) AGCTA-A(6) A a 2012 Poarson,ine Note that four different reactions must be run,one with each dideoxynucleotide
In the manual sequencing method, a single strand of the DNA to be sequenced is mixed with a primer, DNA polymerase I, four dNTP (one of which is radiolabeled), and a small amount of one of the dideoxynucleotides. Note that four different reactions must be run, one with each dideoxynucleotide
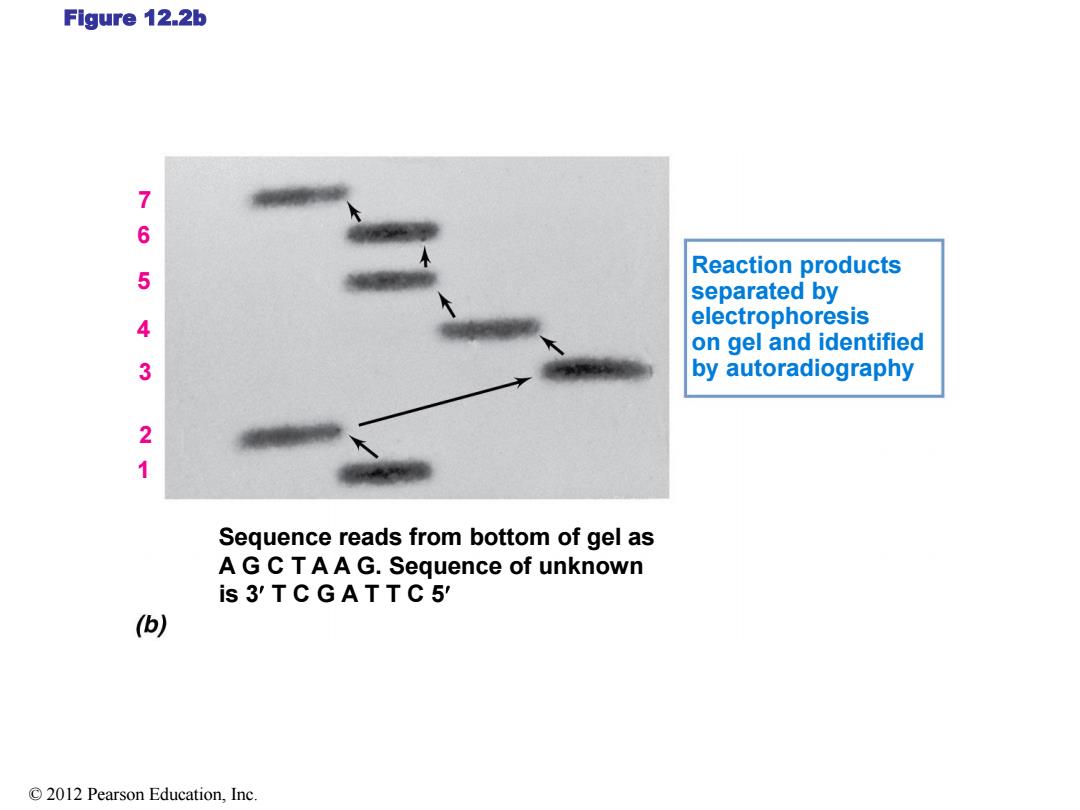
Figure 12.2b 7 6 Reaction products 5 separated by 4 electrophoresis on gel and identified 3 by autoradiography 2 1 Sequence reads from bottom of gel as A G C TAA G.Sequence of unknown is 3'TCGATTC5' () 2012 Pearson Education,Inc
Figure 12.2b Reaction products separated by electrophoresis on gel and identified by autoradiography 7 6 5 4 3 2 1 Sequence reads from bottom of gel as A G C T A A G. Sequence of unknown is 3 T C G A T T C 5 © 2012 Pearson Education, Inc
automated DNA sequencing systems Large-scale sequencing projects have led to automated DNA sequencing systems . Based on Sanger method Radioactivity replaced by fluorescent dye Chen Feng,Lecture of Microbiology
Chen Feng, Lecture of Microbiology automated DNA sequencing systems Large-scale sequencing projects have led to automated DNA sequencing systems • Based on Sanger method • Radioactivity replaced by fluorescent dye