
PREFACE broadenin s has pr er.i al of the beginning graduate students while providing sufficient detail useful to establishe microbial physiologists.This new edi addressing th d not i netics and growth:and genom oloyed b chapters on microbial stress r responses and bacterial differentiation and have added host-paras that corre es microbial physiology wit the field or a p fessor who has been in the field for forty come to be learned. Ie pa
PREFACE The field of microbial physiology has expanded at an incredibly rapid pace since the last edition of this text. The development and implementation of new, highly sophisticated, techniques to study the molecular genetics and physiology of an ever broadening range of microbes has prompted us to write a fourth edition to this book. To give full measure to the extraordinary advances made in microbial physiology we have found it necessary to reorder, separate, and add new material. However, in doing so we have attempted to remain true to the goal of the first edition of “Moat’s Notes” and each subsequent edition by targeting discussions to undergraduate and beginning graduate students while providing sufficient detail useful to established microbial physiologists. This new edition continues the tradition of addressing the physiology of a variety of microbes and not just Escherichia coli. We have updated chapters on bacterial structures, intermediary metabolism, genetics and growth; and added chapters discussing the genomic and proteomic methodologies employed by the new breed of microbial physiologist. We have reorganized, updated and expanded chapters on microbial stress responses and bacterial differentiation and have added a chapter on host-parasite interactions that correlates microbial physiology with microbial pathogenesis. We hope that the reader, be they an advanced undergraduate entering the field or a professor who has been in the field for forty years, will come to better appreciate the elegant simplicities and the intricate complexities of microbial physiology, while at the same time realizing that there is still much to be learned. The authors would like to thank the many students, colleagues, and family who provided help and encouragement as we compiled this new edition. We are particularly xix

XX PREFACE ALBERT G.MOAT JOHN W.FOSTER MICHAEL P.SPECTOR air咖
xx PREFACE thankful to those who granted us permission to use figures or illustrations and/or provided us with original materials for this purpose. ALBERT G. MOAT JOHN W. FOSTER MICHAEL P. SPECTOR Huntington, West Virginia Mobile, Alabama

haoPhaobgbaG.Mdlefr。g2i 0471-39483- CHAPTER 1 INTRODUCTION TO MICROBIAL PHYSIOLOGY THE ESCHERICHIA COLI PARADIGM known on this topic within on book.However.olid foundtion Other organisms th here is to offer a point of conflu is provided CELL STRUCTURE ammbrane-bound nucleus s do eukaryouie mierooranism therefore.theyar prokaryotic.In addition to these basi e types of bact The Cell Surface The interface bety n the microbial cell and its external environm nt is the cell surface It protects the cell interior from extemal hazards and maintains the integrity of the cell
CHAPTER 1 INTRODUCTION TO MICROBIAL PHYSIOLOGY THE ESCHERICHIA COLI PARADIGM Microbial physiology is an enormous discipline encompassing the study of thousands of different microorganisms. It is, of course, foolhardy to try to convey all that is known on this topic within the confines of one book. However, a solid foundation can be built using a limited number of organisms to illustrate key concepts of the field. This text helps set the foundation for further inquiry into microbial physiology and genetics. The gram-negative organism Escherichia coli is used as the paradigm. Other organisms that provide significant counterexamples to the paradigm or alternative strategies to accomplish a similar biochemical goal are also included. In this chapter we paint a broad portrait of the microbial cell with special focus on E. coli. Our objective here is to offer a point of confluence where the student can return periodically to view how one aspect of physiology might relate to another. Detailed treatment of each topic is provided in later chapters. CELL STRUCTURE As any beginning student of microbiology knows, bacteria come in three basic models: spherical (coccus), rod (bacillus), and spiral (spirillum). They do not possess a membrane-bound nucleus as do eukaryotic microorganisms; therefore, they are prokaryotic. In addition to these basic types of bacteria, there are other more specialized forms described as budding, sheathed, and mycelial. Figure 1-1 presents a schematic representation of a typical (meaning E. coli) bacterial cell. The Cell Surface The interface between the microbial cell and its external environment is the cell surface. It protects the cell interior from external hazards and maintains the integrity of the cell 1 Microbial Physiology. Albert G. Moat, John W. Foster and Michael P. Spector Copyright ¶ 2002 by Wiley-Liss, Inc. ISBN: 0-471-39483-1

LPS- Periplasm roteins HO子inner Membrane Flagella Coupled Transcription Translation 00 proteins (ca.108 mo ca.108 molecules/cell) Fig.1-1.Diag matic repr sentation of a "typical"bacterial cell (Escherichia coli). Portions of the cell are enlarged to show further details. as a discrete entity.Although it must be steadfa include carbohydra es (e.g.glucose).vitamins (e.g. vitamin B mino acids.and a exported to the e or the c Cell Wall.In 1 stigator Chr ed a di after dece ation.Cells that retained the stain were very dis the gra vall of e has wo mapo tures:th ell of multiple layers of pepti which is a linea unit N-acetylgl mine nd N-acety rt pep the peptide he cross-b ging b By contrast,th
2 INTRODUCTION TO MICROBIAL PHYSIOLOGY Peptidoglycan Pores } Outer Membrane } } Periplasm Inner Membrane Lipoprotein LPS Phospholipid Proteins Chromosome RNA Coupled Transcription - Translation Polyribosomes RNA polymerase Flagella Cytosol 1000−2000 proteins ( ca. 106 molecules/cell) 60 tRNAs ( ca. 106 molecules/cell) Glycogen Fig. 1-1. Diagrammatic representation of a “typical” bacterial cell (Escherichia coli). Portions of the cell are enlarged to show further details. as a discrete entity. Although it must be steadfast in fulfilling these functions, it must also enable transport of large molecules into and out of the cell. These large molecules include carbohydrates (e.g., glucose), vitamins (e.g., vitamin B12), amino acids, and nucleosides, as well as proteins exported to the exterior of the cell. The structure and composition of different cell surfaces can vary considerably depending on the organism. Cell Wall. In 1884, the Danish investigator Christian Gram devised a differential stain based on the ability of certain bacterial cells to retain the dye crystal violet after decoloration with 95% ethanol. Cells that retained the stain were called gram positive. Subsequent studies have shown that this fortuitous discovery distinguished two fundamentally different types of bacterial cells. The surface of gram-negative cells is much more complex than that of gram-positive cells. As shown in the schematic drawings in Figure 1-2, the gram-positive cell surface has two major structures: the cell wall and the cell membrane. The cell wall of gram-positive cells is composed of multiple layers of peptidoglycan, which is a linear polymer of alternating units of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). A short peptide chain is attached to muramic acid. A common feature in bacterial cell walls is cross-bridging between the peptide chains. In a gram-positive organism such as Staphylococcus aureus, the cross-bridging between adjacent peptides may be close to 100%. By contrast, the frequency of cross-bridging in Escherichia coli (a gram-negative organism) may be as low as 30% (Fig. 1-3). Other components — for example, lipoteichoic acid (only
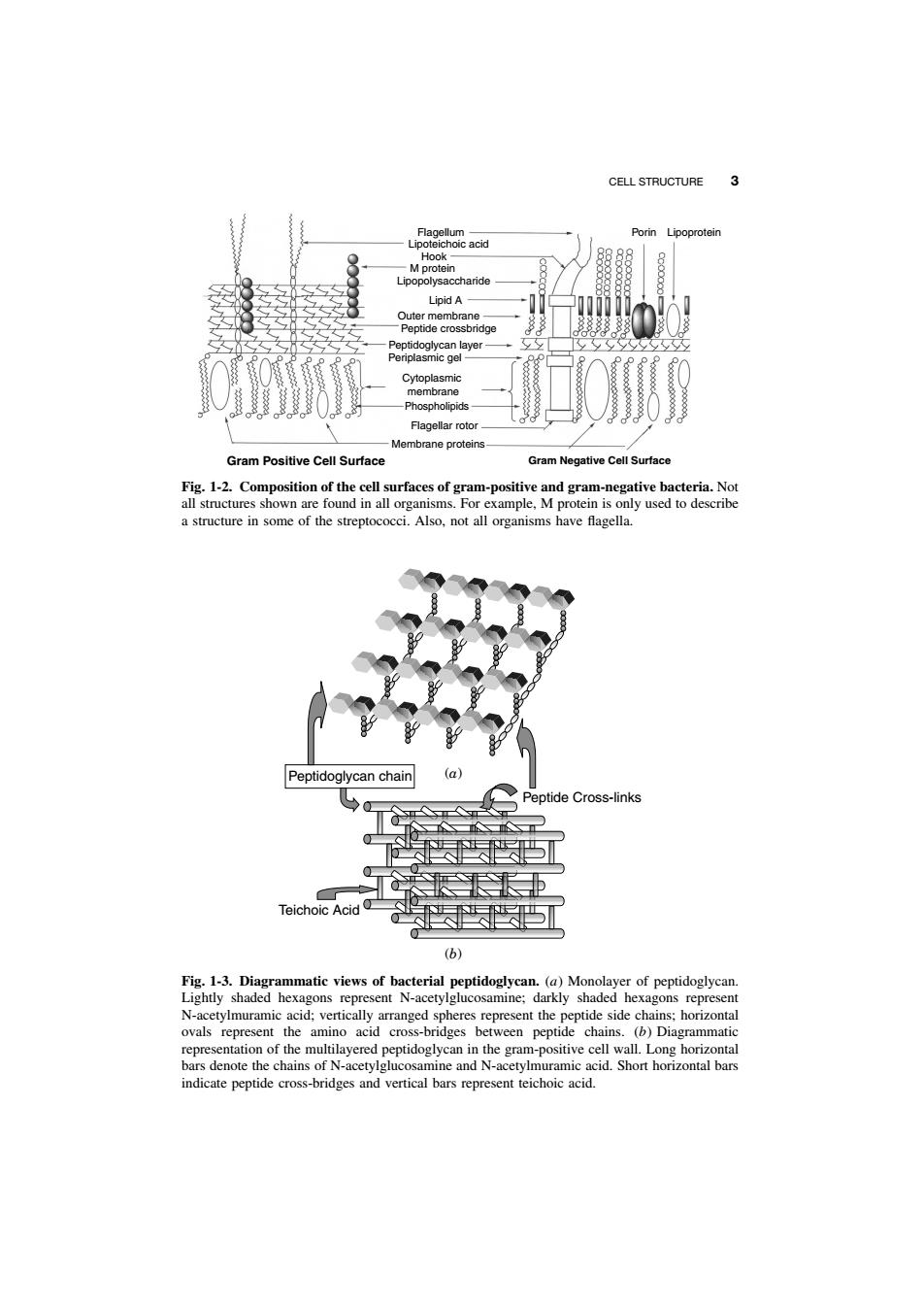
CELL STRUCTURE 3 age0eacd Porin Lipoprote LupA Membrane proteins Gram Positive Cell Surface Gram Negative Cell Surfac a structure in some of the streptococci.Also,not all organisms have flagella. Peptidoglycan chain ptide Cross-link 长P (b) atic views of bacterial an (a)Mon he amino acid s bet en pept ,denote t e and e pep
CELL STRUCTURE 3 Flagellum Lipoteichoic acid Hook M protein Lipopolysaccharide Lipid A Outer membrane Peptide crossbridge Peptidoglycan layer Periplasmic gel Cytoplasmic membrane Phospholipids Flagellar rotor Membrane proteins Porin Lipoprotein Gram Positive Cell Surface Gram Negative Cell Surface Fig. 1-2. Composition of the cell surfaces of gram-positive and gram-negative bacteria. Not all structures shown are found in all organisms. For example, M protein is only used to describe a structure in some of the streptococci. Also, not all organisms have flagella. Peptide Cross-links Teichoic Acid Peptidoglycan chain (b) (a) Fig. 1-3. Diagrammatic views of bacterial peptidoglycan. (a) Monolayer of peptidoglycan. Lightly shaded hexagons represent N-acetylglucosamine; darkly shaded hexagons represent N-acetylmuramic acid; vertically arranged spheres represent the peptide side chains; horizontal ovals represent the amino acid cross-bridges between peptide chains. (b) Diagrammatic representation of the multilayered peptidoglycan in the gram-positive cell wall. Long horizontal bars denote the chains of N-acetylglucosamine and N-acetylmuramic acid. Short horizontal bars indicate peptide cross-bridges and vertical bars represent teichoic acid