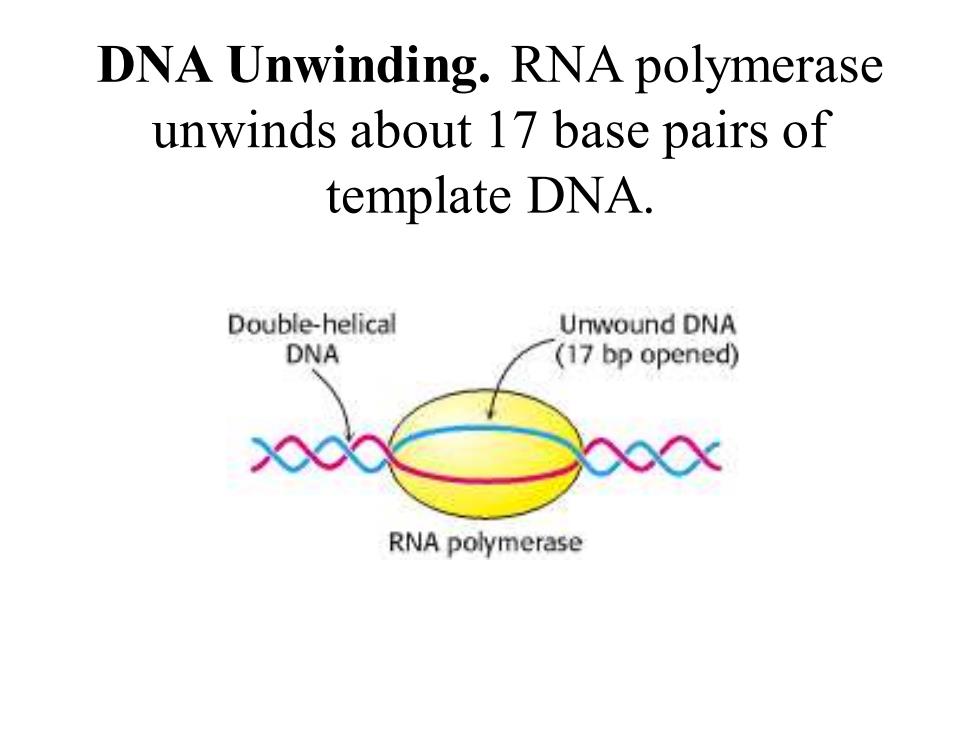
DNA Unwinding.RNA polymerase unwinds about 17 base pairs of template DNA. Double-helical Unwound DNA DNA (17 bp opened) 00x RNA polymerase
DNA Unwinding. RNA polymerase unwinds about 17 base pairs of template DNA
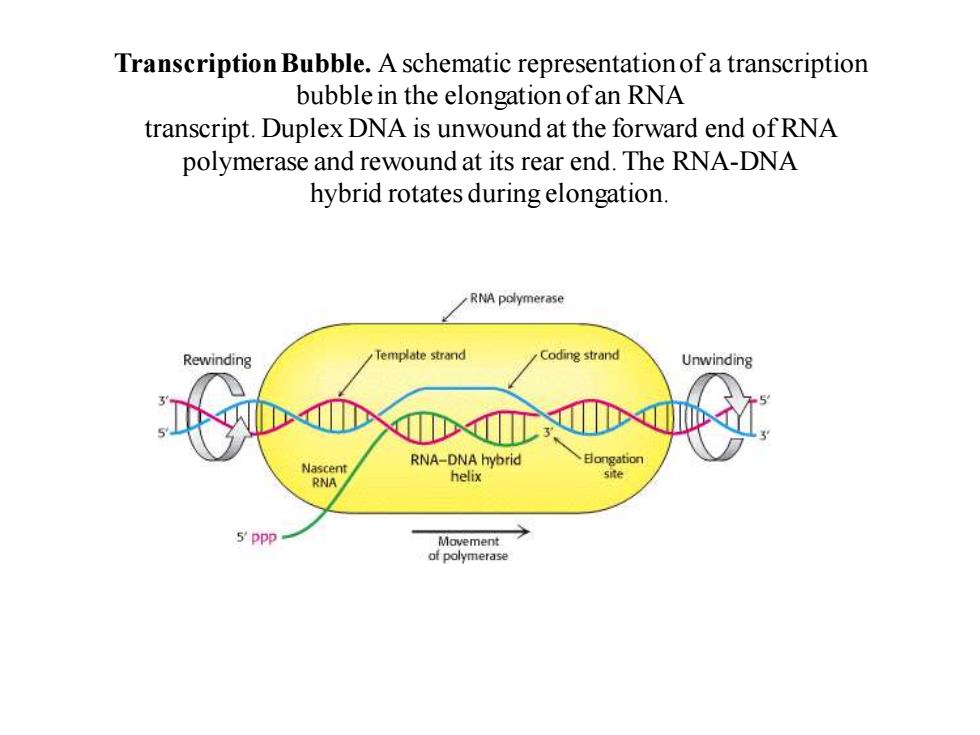
Transcription Bubble.A schematic representation of a transcription bubble in the elongation of an RNA transcript.Duplex DNA is unwound at the forward end of RNA polymerase and rewound at its rear end.The RNA-DNA hybrid rotates during elongation. RNA polymerase Rewinding Template strand Coding strand Unwinding 日ongation Nascent RNA-DNA hybrid RNA helix site 5'ppp Movement of polymerase
Transcription Bubble. A schematic representation of a transcription bubble in the elongation of an RNA transcript. Duplex DNA is unwound at the forward end of RNA polymerase and rewound at its rear end. The RNA-DNA hybrid rotates during elongation

(5)CGCTATAGCGTTT(3) DNA nontemplate(coding)strand (3)GCGATATCGCAAA(5) DNA template strand (5)CGCUAUAGCGUUU(3) RNA transcript
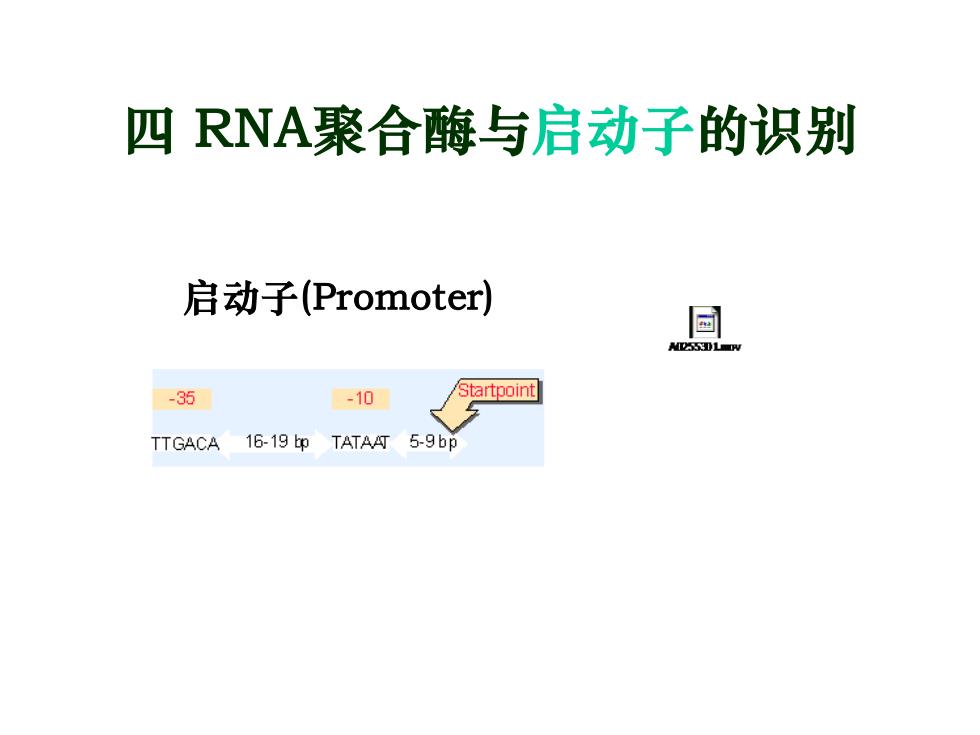
四RNA聚合酶与启动子的识别 启动子(Promoter) -35 -10 Startpoint TTGACA 16-19 bp TATAAT 5-9bp
四 RNA聚合酶与启动子的识别 启动子(Promoter)
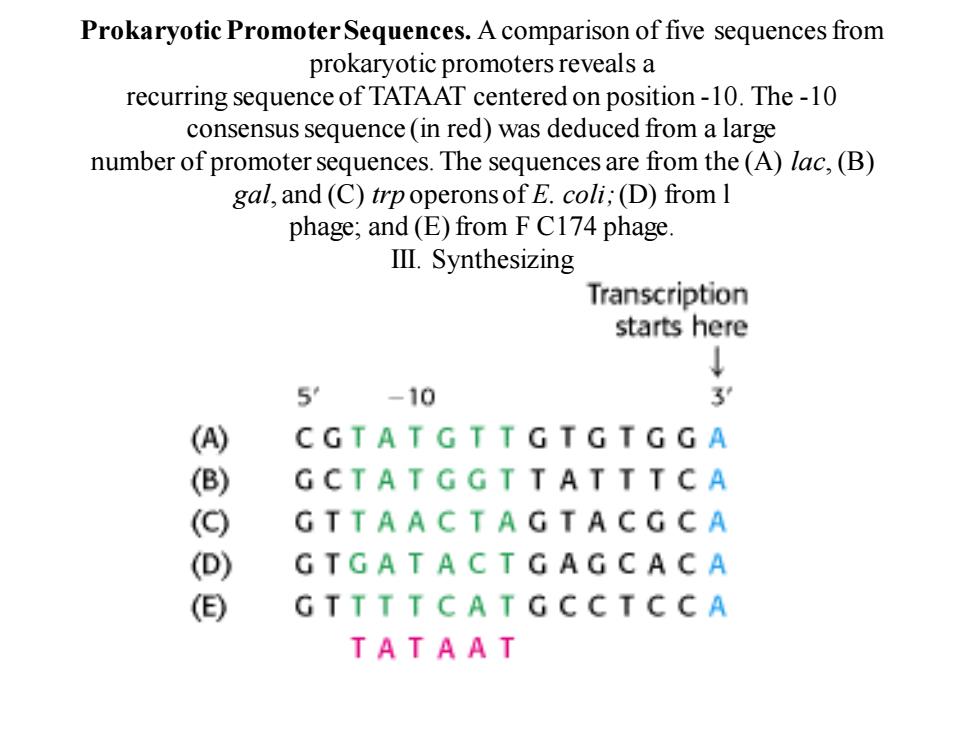
Prokaryotic Promoter Sequences.A comparison of five sequences from prokaryotic promoters reveals a recurring sequence of TATAAT centered on position-10.The-10 consensus sequence (in red)was deduced from a large number of promoter sequences.The sequences are from the(A)lac,(B) gal,and (C)trpoperons of E.coli;(D)from I phage;and (E)from F C174 phage lⅢl.Synthesizing Transcription starts here ↓ 5 -10 3 () CGTATGTTGTGTGGA (B) GCTATGGTTATTTCA (G) GTTAACTAGTACGCA (D) GTGATACT GAGCACA () GTTTTCATGCC T CCA TATAAT
Prokaryotic Promoter Sequences. A comparison of five sequences from prokaryotic promoters reveals a recurring sequence of TATAAT centered on position -10. The -10 consensus sequence (in red) was deduced from a large number of promoter sequences. The sequences are from the (A) lac, (B) gal, and (C) trp operons of E. coli; (D) from l phage; and (E) from F C174 phage. III. Synthesizing