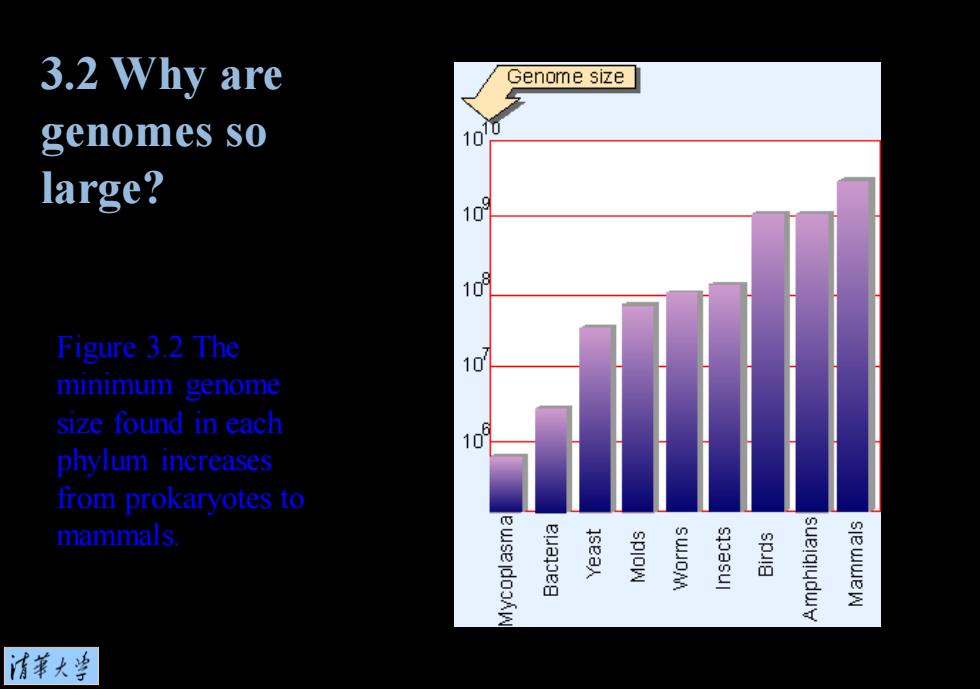
3.2 Why are Genome size genomes so 0 10 large? 109 109 Figure 3.2 The 10 minimum genome size found in each 10 phylum increases from prokaryotes to mammals ewse jdooAw sea spIoW s1oasul spulg 清菜大当
Figure 3.2 The minimum genome size found in each phylum increases from prokaryotes to mammals. 3.2 Why are genomes so large?

3.2 Why are genomes so large? Phylum Species Genome (bp) Algae Pyrenomas salina 6.6×105 Mycoplasma M.pneumoniae 1.0X19 Bacterium E.coli 4.2x109 Yeast S. cerevisiae 1.3×10 Slime mold D.discoideum 5.4×107 Nem atode C.elegans 8.0×107 Insect D.melanogaster 1.4× 108 Bird G.domesticus 1.2x 109 Amphibian X.laevis 3.1×109 Mammal H. sapiens 3.3× 109 Figure 3.3 The genome sizes of some common experimental animals 情菜大当
Figure 3.3 The genome sizes of some common experimental animals. 3.2 Why are genomes so large?
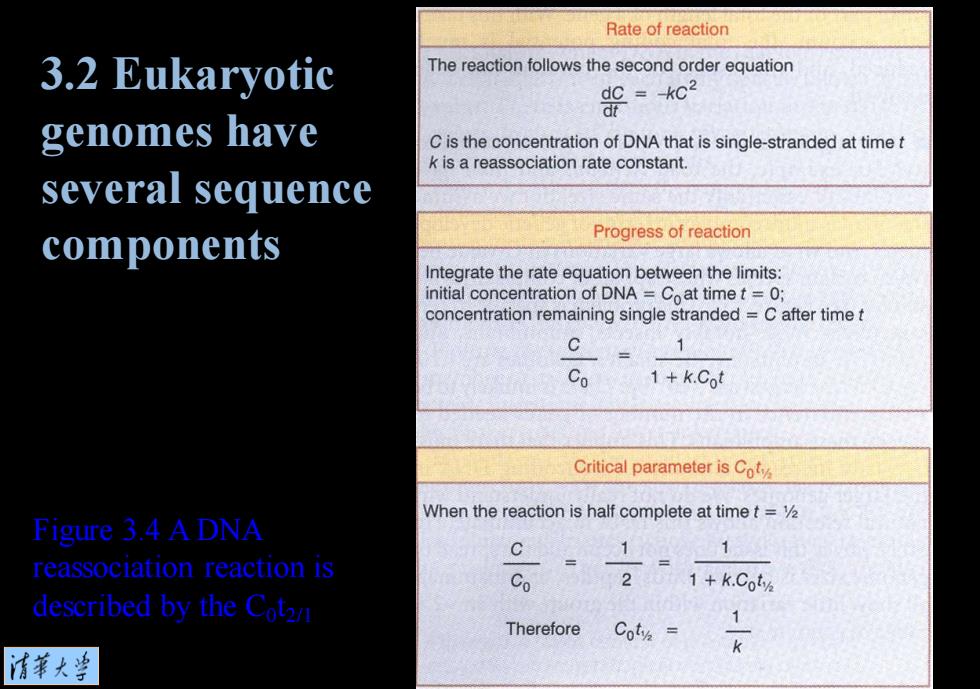
Rate of reaction 3.2 Eukaryotic The reaction follows the second order equation =-02 dt genomes have C is the concentration of DNA that is single-stranded at time t k is a reassociation rate constant. several sequence components Progress of reaction Integrate the rate equation between the limits: initial concentration of DNA =Coat time t=0; concentration remaining single stranded=C after time t 1 Co 1+k.Cot Critical parameter is Coty When the reaction is half complete at time t=V Figure 3.4 A DNA C 1 1 reassociation reaction is Co 2 1+k.Coty described by the Cotzn Therefore 1 Coty= 清菜大当
Figure 3.4 A DNA reassociation reaction is described by the C0 t2/1 3.2 Eukaryotic genomes have several sequence components
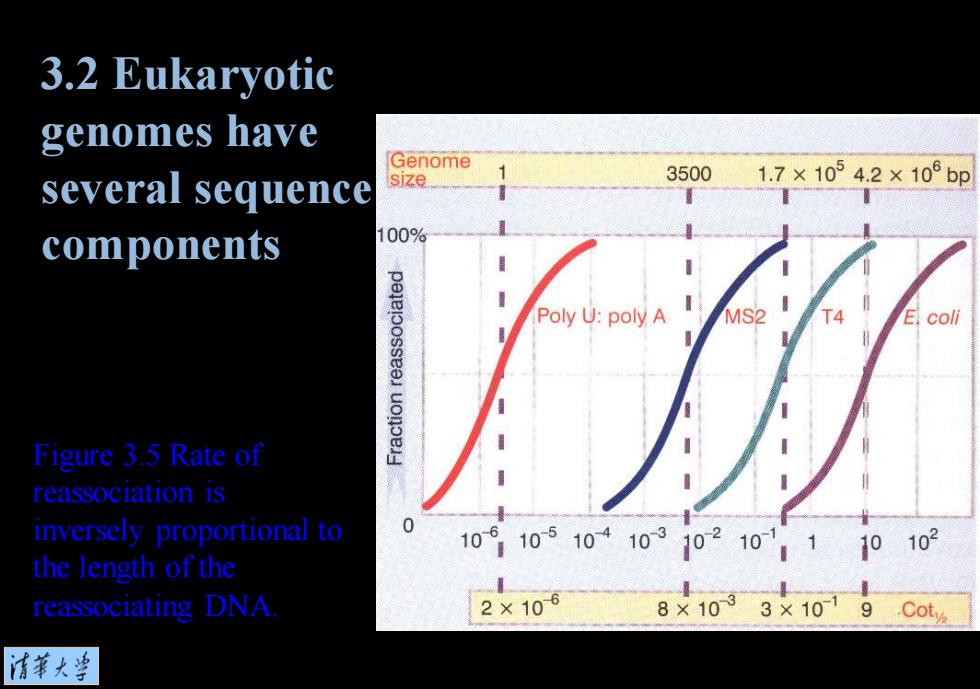
3.2 Eukaryotic genomes have Genome several sequence 35001.7×10°4.2×10°bp components 100% Poly U:poly A MS2 T4 E.coli Figure 3.5 Rate of reassociation is inversely proportional to 10910510410310210110102 the length of the reassociating DNA 2×106 8×1033×1019 .Cot 情菜大当
Figure 3.5 Rate of reassociation is inversely proportional to the length of the reassociating DNA. 3.2 Eukaryotic genomes have several sequence components
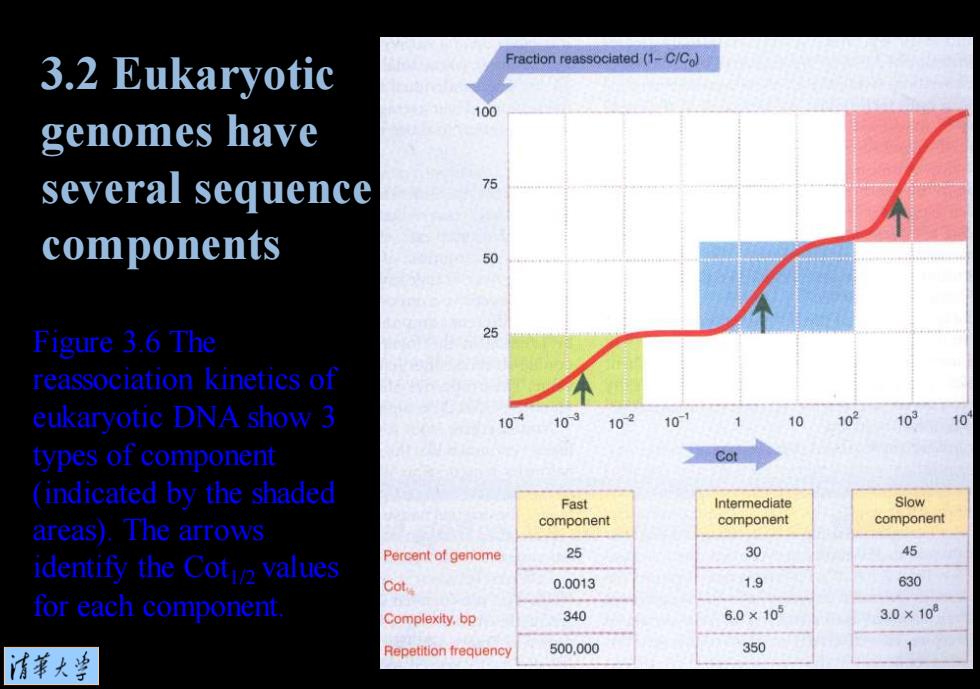
3.2 Eukaryotic Fraction reassociated(1-C/Co) 100 genomes have several sequence 75 components 50 Figure 3.6 The 25 reassociation kinetics of eukaryotic DNA show 3 10 10 102 10 10 10 103 10 types of component Cot (indicated by the shaded Fast Intermediate Slow areas).The arrows component component component Percent of genome 25 30 45 identify the Cot values 0.0013 1.9 630 for each component Complexity,bp 340 6.0×105 3.0×10 Repetition frequency 500.000 350 1 清菜大当
Figure 3.6 The reassociation kinetics of eukaryotic DNA show 3 types of component (indicated by the shaded areas). The arrows identify the Cot1/2 values for each component. 3.2 Eukaryotic genomes have several sequence components