bZIP and bHLH Motifs A number of transcription factors have a highly basic DNA-binding motif linked to protein dimerization motifs -Leucine zippers -Helix-loop-helix Examples include: -CCAAT/enhancer-binding protein -MyoD protein 12-6
12-6 bZIP and bHLH Motifs ✓ A number of transcription factors have a highly basic DNA-binding motif linked to protein dimerization motifs – Leucine zippers – Helix-loop-helix ✓Examples include: – CCAAT/enhancer-binding protein – MyoD protein
Transcription-Activating Domains Most activators have one of these domains, Some have more than one These domains fall into three classes: - Acidic domains(酸性域)such as yeast GAL4 with 11 acidic amino acids out of 49 amino acids in the domain Glutamine-rich domains(谷酰胺富含域) include Sp1 having 2 that are 25%glutamine Proline-rich domains(脯氨酸富含域)such as CTF which has a domain of 84 amino acids, 19 proline 12-7
12-7 Transcription-Activating Domains ✓ Most activators have one of these domains, Some have more than one ✓ These domains fall into three classes: – Acidic domains (酸性域) such as yeast GAL4 with 11 acidic amino acids out of 49 amino acids in the domain – Glutamine-rich domains (谷酰胺富含域) include Sp1 having 2 that are 25% glutamine – Proline-rich domains (脯氨酸富含域) such as CTF which has a domain of 84 amino acids, 19 proline
12.2 Structures of the DNA-Binding Motifs of Activators DNA-binding domains have well-defined structures √X-ray crystallographic(X射线晶体衍射)studies have shown how these structures interact with their DNA targets √Interaction domains forming dimers(二聚体), or tetramers(四聚体),have also been described Most classes of DNA-binding proteins can't bind DNA in monomer form 12-8
12-8 12.2 Structures of the DNA-Binding Motifs of Activators ✓ DNA-binding domains have well-defined structures ✓ X-ray crystallographic (X射线晶体衍射) studies have shown how these structures interact with their DNA targets ✓ Interaction domains forming dimers (二聚体), or tetramers (四聚体), have also been described ✓ Most classes of DNA-binding proteins can’t bind DNA in monomer form

Zinc Fingers Described by Klug in TFIIIA Nine repeats of a 30-residue element: -2 closely spaced cysteines(半胱氦 followed 12 amino acids later by 2 closely spaced histidines(组氨酸) -Rich in zinc,enough for 1 zinc ion per repeat -Each zinc ion is complexed by 2 cysteines and 2 histidines in each repeat to form the finger-shaped structure or domain -Specific recognition between the zinc finger and its DNA target occurs in the major groove 12-9
12-9 Zinc Fingers ✓Described by Klug in TFIIIA ✓Nine repeats of a 30-residue element: –2 closely spaced cysteines (半胱氨 酸) followed 12 amino acids later by 2 closely spaced histidines (组氨酸) –Rich in zinc, enough for 1 zinc ion per repeat –Each zinc ion is complexed by 2 cysteines and 2 histidines in each repeat to form the finger-shaped structure or domain –Specific recognition between the zinc finger and its DNA target occurs in the major groove Zn
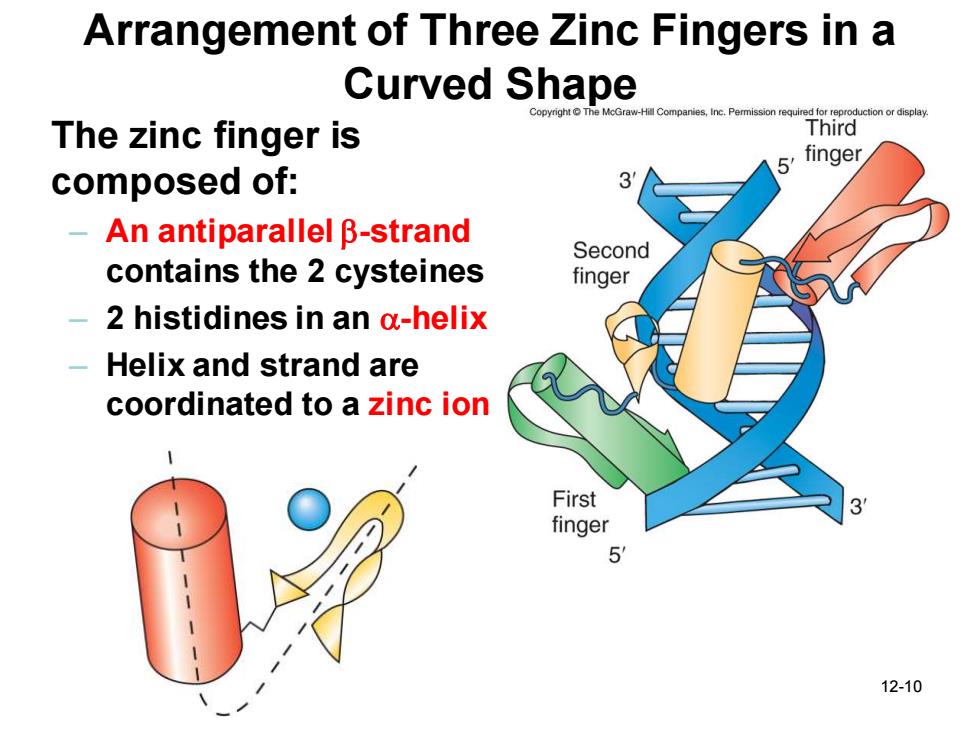
Arrangement of Three Zinc Fingers in a Curved Shape he McGraw-Hill Companies,Inc.Permission required fo roduction or display The zinc finger is Third finger composed of: An antiparallel B-strand Second contains the 2 cysteines finger 2 histidines in an a-helix Helix and strand are coordinated to a zinc ion First finger 6 12-10
12-10 Arrangement of Three Zinc Fingers in a Curved Shape The zinc finger is composed of: – An antiparallel b-strand contains the 2 cysteines – 2 histidines in an a-helix – Helix and strand are coordinated to a zinc ion