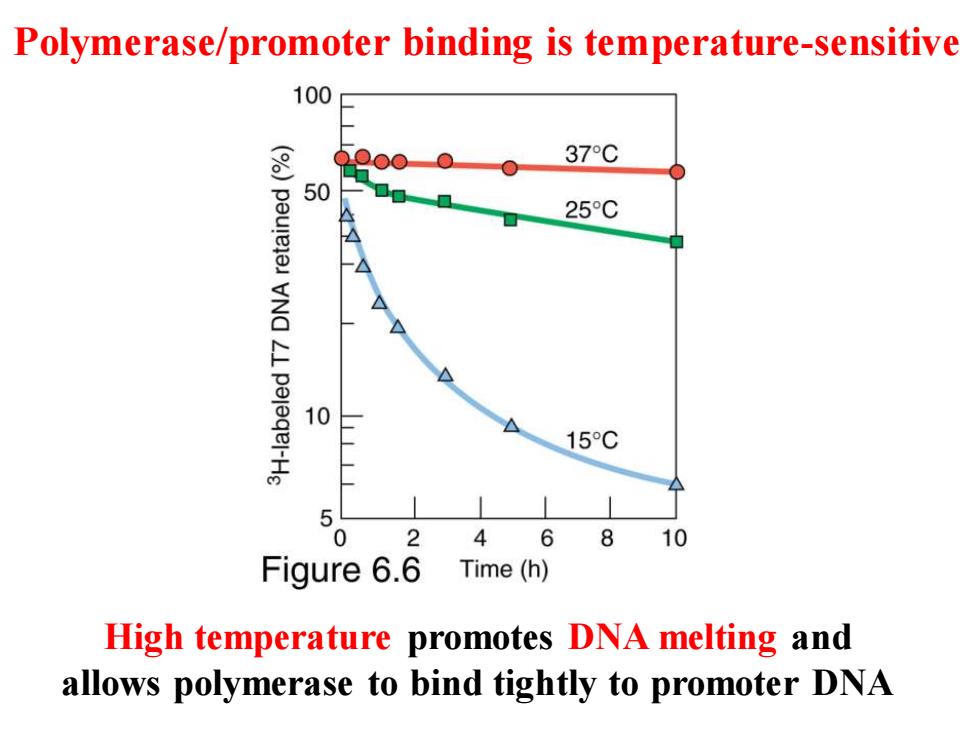
Polymerase/promoter binding is temperature-sensitive 100 37°C 50 25C 10 15C 5 0 4 6810 Figure 6.6 Time(h) High temperature promotes DNA melting and allows polymerase to bind tightly to promoter DNA
Polymerase/promoter binding is temperature-sensitive High temperature promotes DNA melting and allows polymerase to bind tightly to promoter DNA
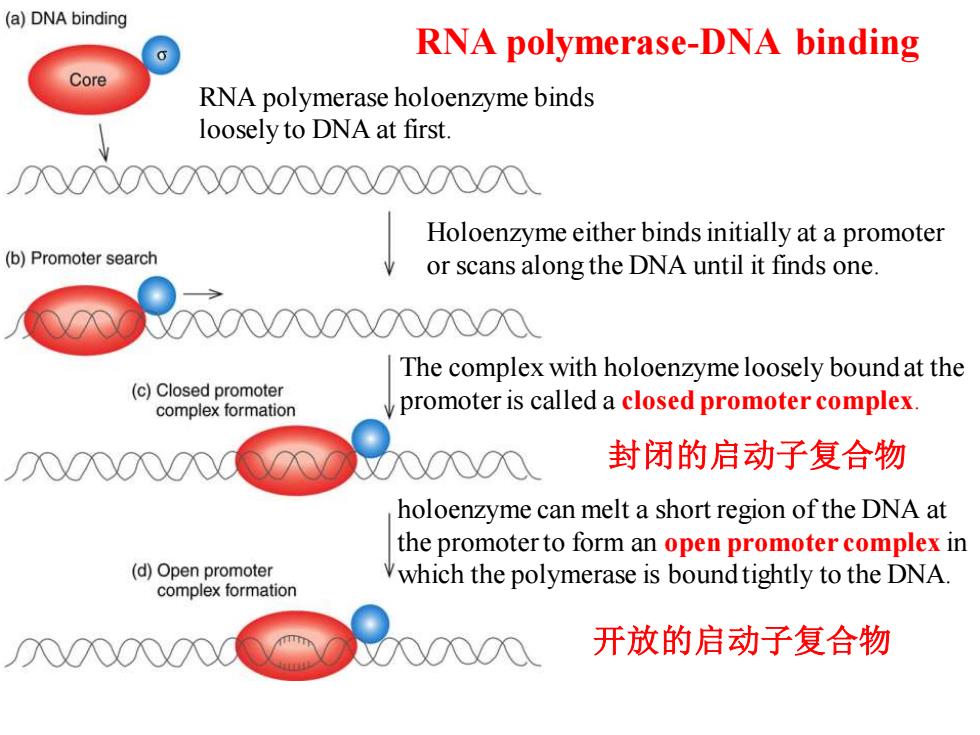
(a)DNA binding RNA polymerase-DNA binding Core RNA polymerase holoenzyme binds loosely to DNA at first. 00000000N000000 Holoenzyme either binds initially at a promoter (b)Promoter search or scans along the DNA until it finds one. The complex with holoenzyme loosely bound at the (c)Closed promoter complex formation promoter is called a closed promoter complex 封闭的启动子复合物 holoenzyme can melt a short region of the DNA at the promoter to form an open promoter complex in (d)Open promoter which the polymerase is bound tightly to the DNA. complex formation 开放的启动子复合物
holoenzyme can melt a short region of the DNA at the promoter to form an open promoter complex in which the polymerase is bound tightly to the DNA. RNA polymerase-DNA binding RNA polymerase holoenzyme binds loosely to DNA at first. The complex with holoenzyme loosely bound at the promoter is called a closed promoter complex. 封闭的启动子复合物 开放的启动子复合物 Holoenzyme either binds initially at a promoter or scans along the DNA until it finds one
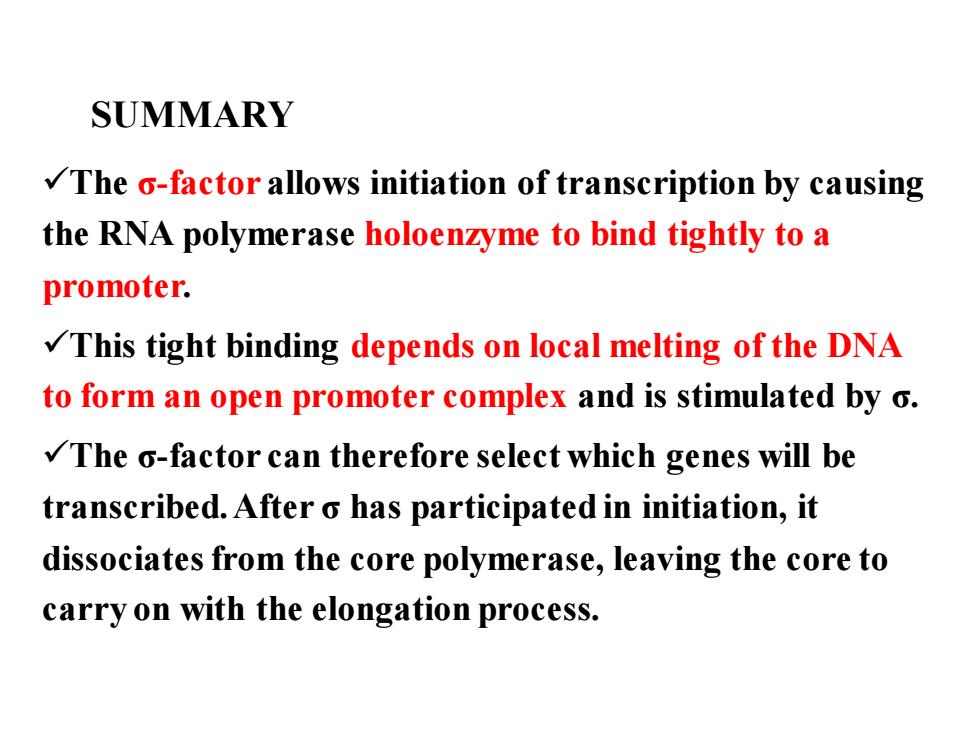
SUMMARY The o-factor allows initiation of transcription by causing the RNA polymerase holoenzyme to bind tightly to a promoter. This tight binding depends on local melting of the DNA to form an open promoter complex and is stimulated by o. The o-factor can therefore select which genes will be transcribed.After o has participated in initiation,it dissociates from the core polymerase,leaving the core to carry on with the elongation process
SUMMARY ✓The σ-factor allows initiation of transcription by causing the RNA polymerase holoenzyme to bind tightly to a promoter. ✓This tight binding depends on local melting of the DNA to form an open promoter complex and is stimulated by σ. ✓The σ-factor can therefore select which genes will be transcribed. After σ has participated in initiation, it dissociates from the core polymerase, leaving the core to carry on with the elongation process
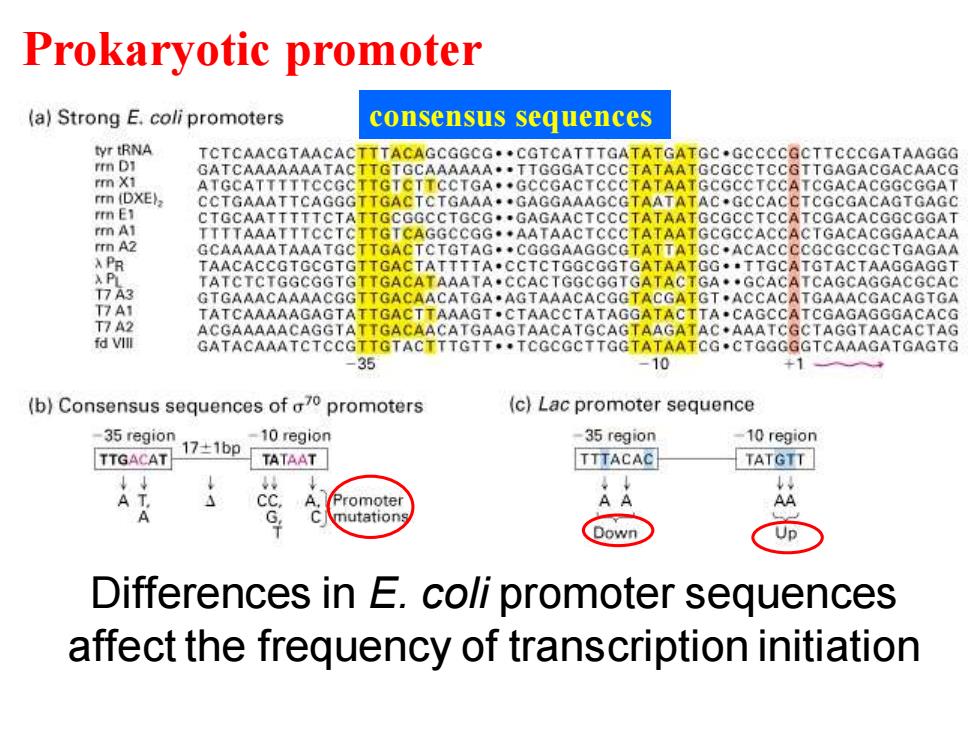
Prokaryotic promoter (a)Strong E.coli promoters consensus sequences tyr tRNA TCTCAACGTAACACTTTACAGCGGCG.CGTCATTTGATATGATGC.GCCCCGCTTCCCGATAAGGG rmn DI GATCAAAAAAATACTTGTGCAAAAAA.TTGGGATCCCTATAATGCGCCTCCGTTGAGACGACAACG rm X1 ATGCATTTTTCCGCTTGTCTTCCTGA.GCCGACTCCCTATAATGCGCCTCCATCGACACGGCGGAT rrn (DXE), CCTGAAATTCAGGGTTGACTCTGAAA.GAGGAAAGCGTAATATAC.GCCACCTCGCGACAGTGAGC rmn E1 CTGCAATTTTTCTATTGCGGCCTGCG.GAGAACTCCCTATAATGCGCCTCCATCGACACGGCGGAT rm A1 TTTTAAATTTCCTCTTGTCAGGCCGG.AATAACTCCCTATAATGCGCCACCACTGACACGGAACAA rrn A2 GCAAAAATAAATGCTTGACTCTGTAG.CGGGAAGGCGTATTATGC.ACACCCCGCGCCGCTGAGAA X PR TAACACCGTGCGTGTTGACTATTTTA.CCTCTGGCGGTGATAATGG.TTGCATGTACTAAGGAGGT XPL TATCTCTGGCGGTGTTGACATAAATA.CCACTGGCGGTGATACTGA.GCACATCAGCAGGACGCAC T7A3 GTGAAACAAAACGGTTGACAACATGA.AGTAAACACGGTACGATGT.ACCACATGAAACGACAGTGA T7A1 ATCAAAAAGAGTATTGACTTAAAGT.CTAACCTATAGGATACTTA.CAGCCATCGAGAGGGACACG T7 A2 ACGAAAAACAGGTATTGACAACATGAAGTAACATGCAGTAAGATAC.AAATCGCTAGGTAACACTAG fd Vlll GATACAAATCTCCGTTGTACTTTGTT.TCGCGCTTGGTATAATCG.CTGGGGGTCAAAGATGAGTG -35 -10 +1 (b)Consensus sequences of a70 promoters (c)Lac promoter sequence -35 region 17±1bp -10 region 35 region -10 region TTGACAT TATAAT TTTACAC TATGTT Promoter G mutation Down Differences in E.coli promoter sequences affect the frequency of transcription initiation
Differences in E. coli promoter sequences affect the frequency of transcription initiation Prokaryotic promoter consensus sequences
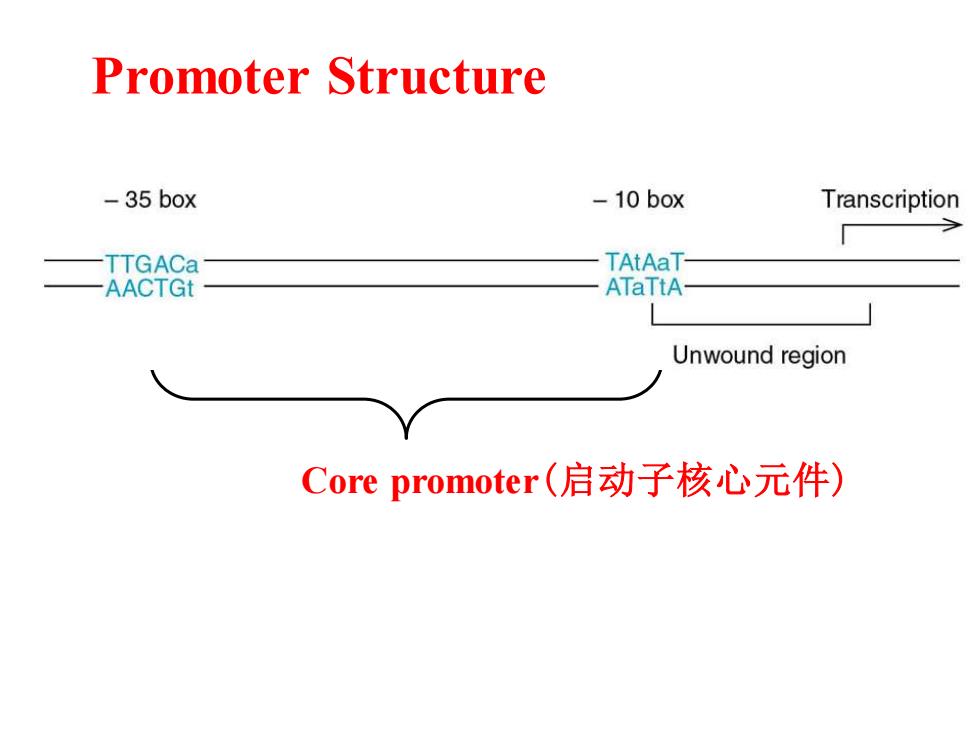
Promoter Structure -35 box -10 box Transcription TTGACa TAtAaT- AACTGt ATaTtA Unwound region Core promoter(启动子核心元件)
Core promoter(启动子核心元件) Promoter Structure