
MD in materials studio Key Modules > Discover:Molecular mechanics computing engine in MS.Based on the carefully deduced force field,the lowest energy configuration,the structure and dynamic trajectory of the molecular system can be accurately calculated by using a variety of molecular mechanics and kinetics methods. > Forcite:Advanced molecular mechanics and dynamics tools.Can be used for fast energy calculation and reliable geometric optimization of molecules or periodic systems. Involved Modules Materials Visualizer:It provides all the tools needed to build structural models of molecules,crystals and polymers.It can operate,observe and analyze structural models,process data in the form of charts,tables or texts,and provide the basic environment and analysis tools for MS.It is the core module of MS. >Amorphous Cell:Constructing complex amorphous models and predicting key properties,usually used with Discovel,Forcite
MD in Materials Studio Key Modules Discover: Molecular mechanics computing engine in MS. Based on the carefully deduced force field, the lowest energy configuration, the structure and dynamic trajectory of the molecular system can be accurately calculated by using a variety of molecular mechanics and kinetics methods. Forcite: Advanced molecular mechanics and dynamics tools. Can be used for fast energy calculation and reliable geometric optimization of molecules or periodic systems. Involved Modules Materials Visualizer: It provides all the tools needed to build structural models of molecules, crystals and polymers. It can operate, observe and analyze structural models, process data in the form of charts, tables or texts, and provide the basic environment and analysis tools for MS. It is the core module of MS. Amorphous Cell: Constructing complex amorphous models and predicting key properties, usually used with Discovel, Forcite
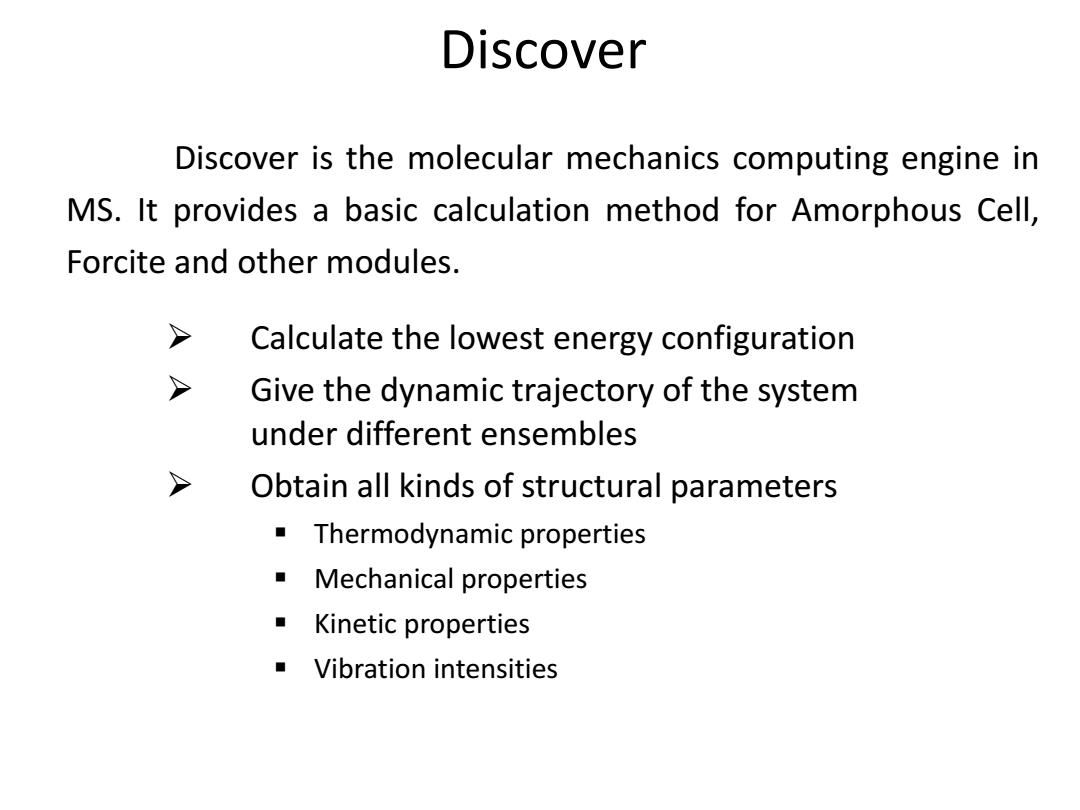
Discover Discover is the molecular mechanics computing engine in MS.It provides a basic calculation method for Amorphous Cell, Forcite and other modules. > Calculate the lowest energy configuration > Give the dynamic trajectory of the system under different ensembles > Obtain all kinds of structural parameters Thermodynamic properties Mechanical properties Kinetic properties Vibration intensities
Discover is the molecular mechanics computing engine in MS. It provides a basic calculation method for Amorphous Cell, Forcite and other modules. Calculate the lowest energy configuration Give the dynamic trajectory of the system under different ensembles Obtain all kinds of structural parameters Thermodynamic properties Mechanical properties Kinetic properties Vibration intensities Discover
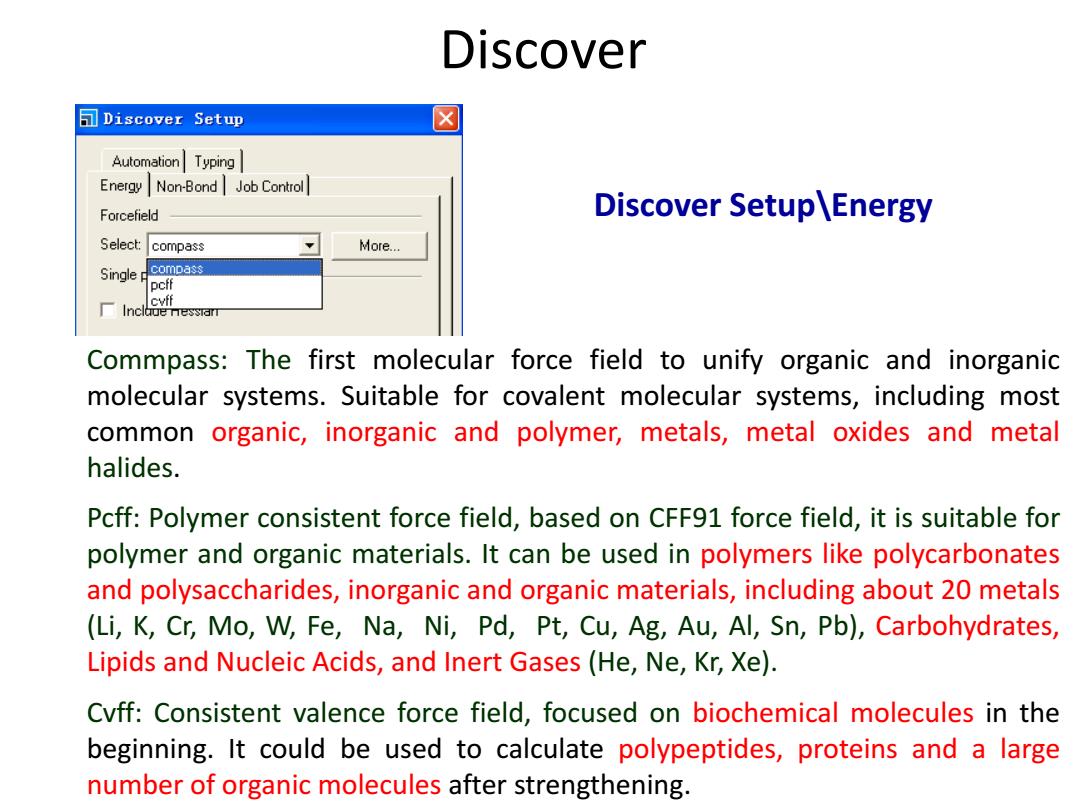
Discover □Discover Setup ☒ Automation Typing Energy Non-Bond Job Control Forcefield Discover Setup\Energy Select: compass Moe… Single compass pcff cvff Inclaue nessian Commpass:The first molecular force field to unify organic and inorganic molecular systems.Suitable for covalent molecular systems,including most common organic,inorganic and polymer,metals,metal oxides and metal halides. Pcff:Polymer consistent force field,based on CFF91 force field,it is suitable for polymer and organic materials.It can be used in polymers like polycarbonates and polysaccharides,inorganic and organic materials,including about 20 metals (Li,K,Cr,Mo,W,Fe,Na,Ni,Pd,Pt,Cu,Ag,Au,Al,Sn,Pb),Carbohydrates, Lipids and Nucleic Acids,and Inert Gases (He,Ne,Kr,Xe). Cvff:Consistent valence force field,focused on biochemical molecules in the beginning.It could be used to calculate polypeptides,proteins and a large number of organic molecules after strengthening
Discover Setup\Energy Commpass: The first molecular force field to unify organic and inorganic molecular systems. Suitable for covalent molecular systems, including most common organic, inorganic and polymer, metals, metal oxides and metal halides. Pcff: Polymer consistent force field, based on CFF91 force field, it is suitable for polymer and organic materials. It can be used in polymers like polycarbonates and polysaccharides, inorganic and organic materials, including about 20 metals (Li, K, Cr, Mo, W, Fe, Na, Ni, Pd, Pt, Cu, Ag, Au, Al, Sn, Pb), Carbohydrates, Lipids and Nucleic Acids, and Inert Gases (He, Ne, Kr, Xe). Cvff: Consistent valence force field, focused on biochemical molecules in the beginning. It could be used to calculate polypeptides, proteins and a large number of organic molecules after strengthening. Discover
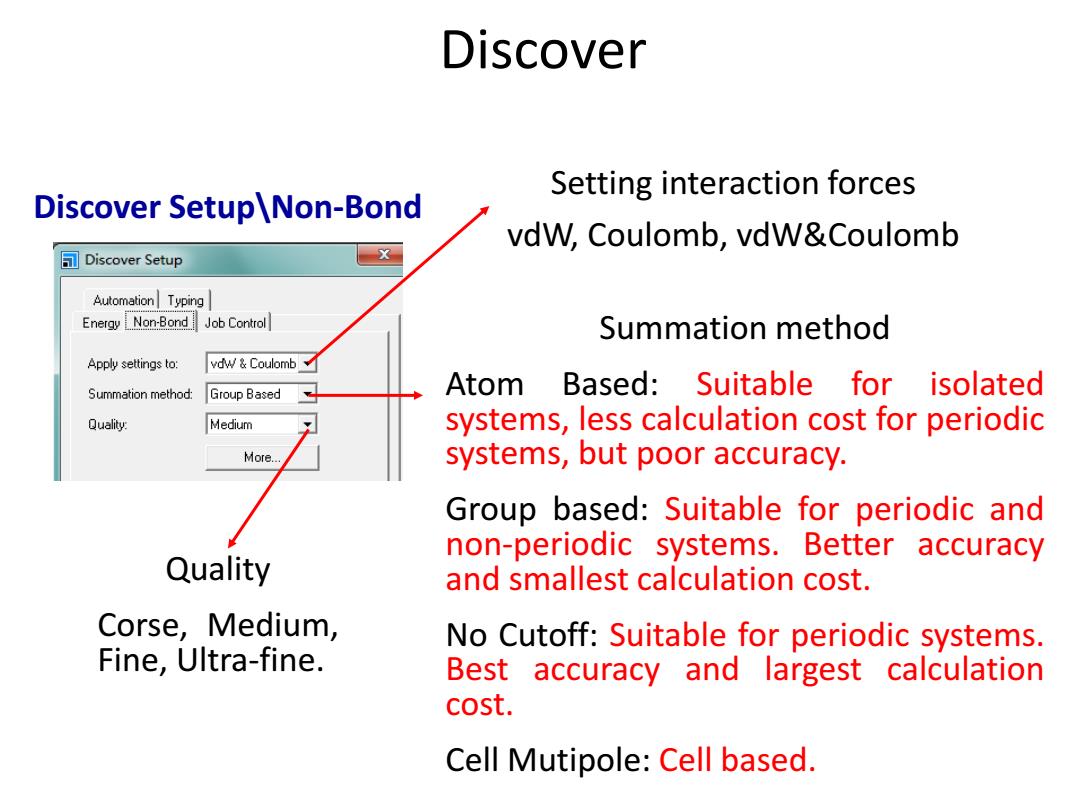
Discover Setting interaction forces Discover Setup\Non-Bond vdW,Coulomb,vdW&Coulomb Discover Setup Automation Typing Energy Non-Bond Job Control Summation method Apply settings to: vdw Coulomb Summation method: Group Based Atom Based:Suitable for isolated Quality: Medium systems,less calculation cost for periodic More.. systems,but poor accuracy. Group based:Suitable for periodic and non-periodic systems.Better accuracy Quality and smallest calculation cost. Corse,Medium, No Cutoff:Suitable for periodic systems. Fine,Ultra-fine. Best accuracy and largest calculation cost. Cell Mutipole:Cell based
Setting interaction forces vdW, Coulomb, vdW&Coulomb Summation method Atom Based: Suitable for isolated systems, less calculation cost for periodic systems, but poor accuracy. Group based: Suitable for periodic and non-periodic systems. Better accuracy and smallest calculation cost. No Cutoff: Suitable for periodic systems. Best accuracy and largest calculation cost. Cell Mutipole: Cell based. Quality Corse, Medium, Fine, Ultra-fine. Discover Discover Setup\Non-Bond
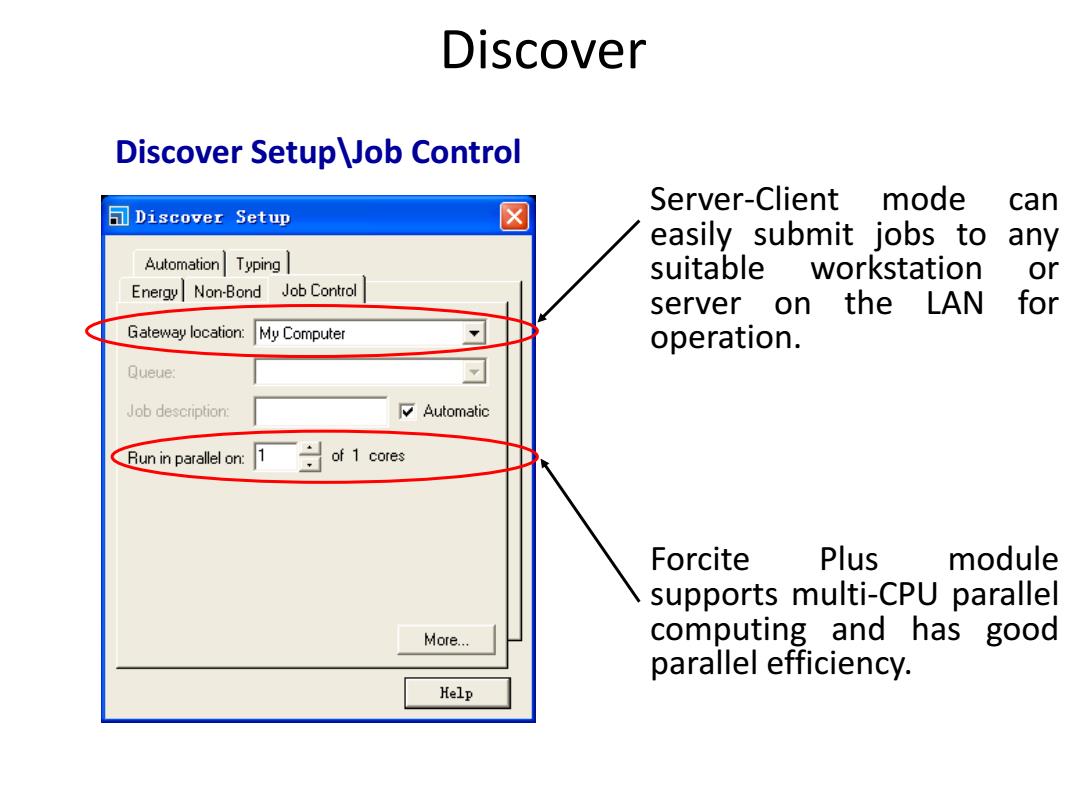
Discover Discover Setup Job Control Server-Client mode can Discover Setup easily submit jobs to any Automation Typing suitable workstation or Energy Non-Bond Job Control server on the LAN for Gateway location: My Computer operation. Queue: Job description: Automatic Run in parallel on: 1 of 1 cores Forcite Plus module supports multi-CPU parallel More… computing and has good parallel efficiency. Kelp
Discover Discover Setup\Job Control Server-Client mode can easily submit jobs to any suitable workstation or server on the LAN for operation. Forcite Plus module supports multi-CPU parallel computing and has good parallel efficiency