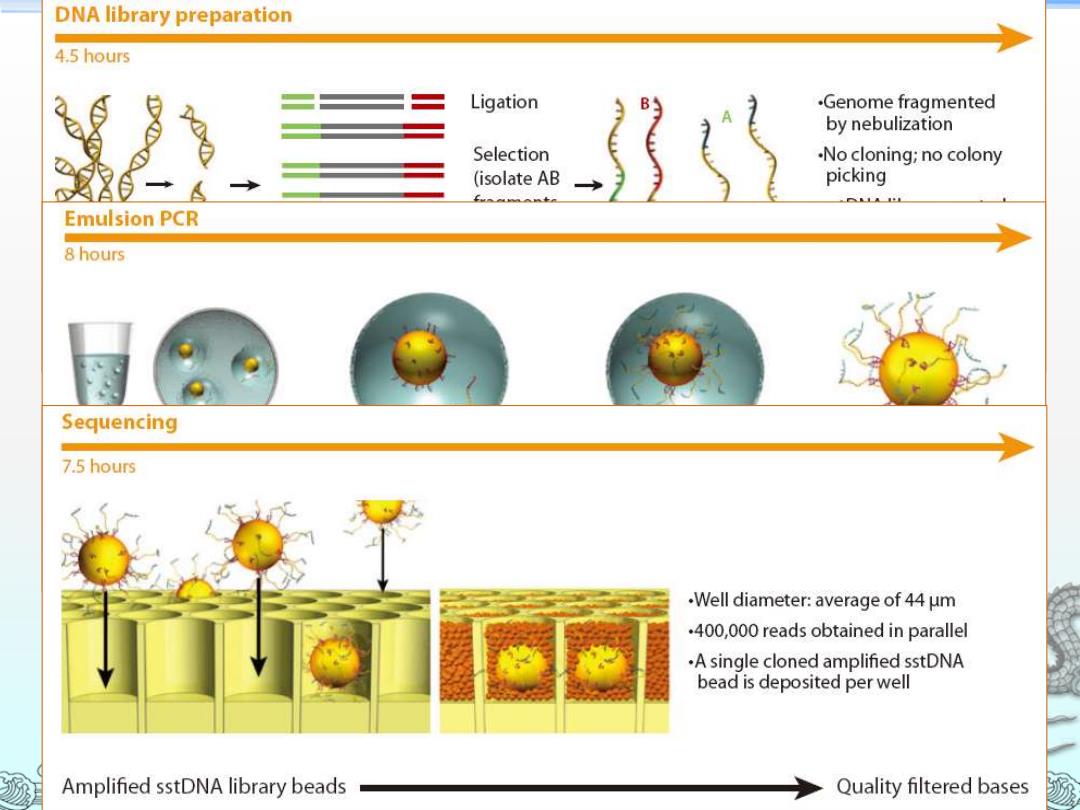
DNA library preparation 4.5 hours Ligation .Genome fragmented by nebulization Selection .No cloning;no colony (isolate AB picking Emulsion PCR 8 hours Sequencing 7.5 hours .Well diameter:average of 44 um .400,000 reads obtained in parallel .A single cloned amplified sstDNA bead is deposited per well Amplified sstDNA library beads Quality filtered bases
Roche/454 FLX Pyrosequencer
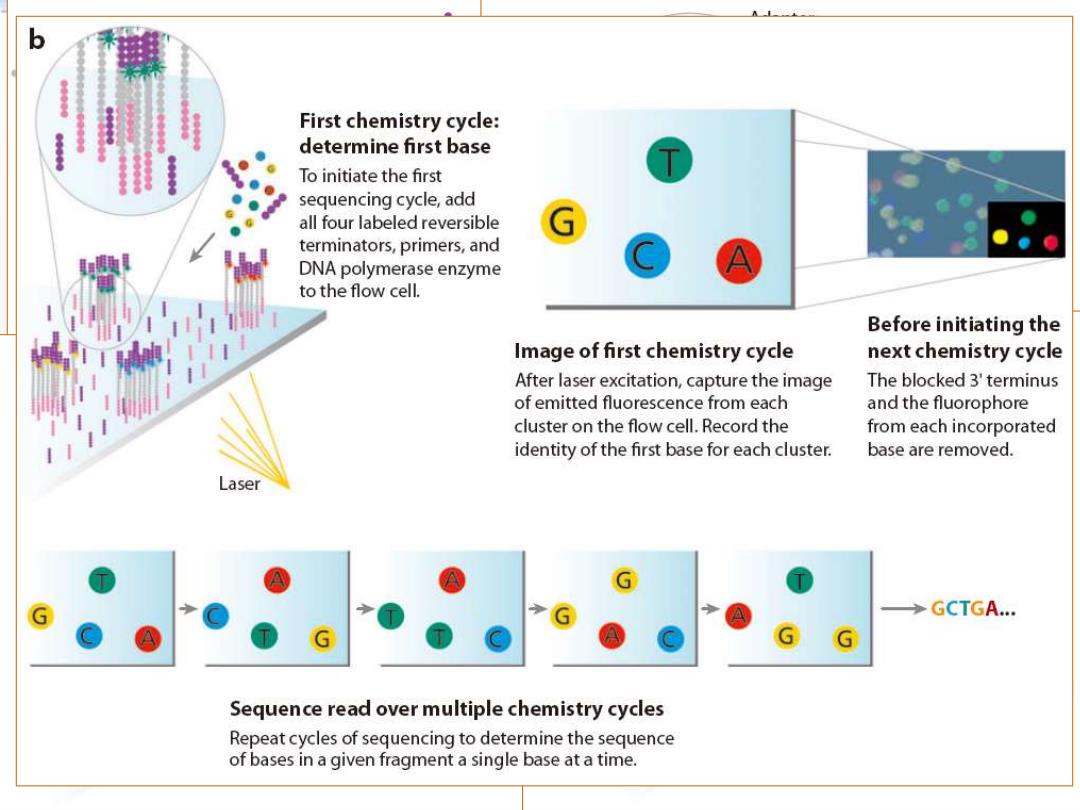
First chemistry cycle: determine first base To initiate the first sequencing cycle,add all four labeled reversible terminators,primers,and DNA polymerase enzyme to the flow cell. Before initiating the Image of first chemistry cycle next chemistry cycle After laser excitation,capture the image The blocked 3'terminus of emitted fluorescence from each and the fluorophore cluster on the flow cell.Record the from each incorporated identity of the first base for each cluster. base are removed. Laser GCTGA... A Sequence read over multiple chemistry cycles Repeat cycles of sequencing to determine the sequence of bases in a given fragment a single base at a time
Illumina Genome Analyzer

8.Repeat Reset with,n-2,n-3,n-4 primers Read position o1234s67so23141shd7lh8l12o2222242s222o到24 1 Universal seq primer (n) 3' Possible dinucleotides encoded by each color puno 2 Universal seq primer(n-1) 2nd base 3' Template sequence A C G 3 Universal seq primer(n-2) 3'm Bridge probe ●● 4 Universal seq primer (n-3) Bridge probe AT AC AA GA 3'i CA CC TC 5 Universal seq primer (n-4) C GT GG ● 3' Bridge probe TG TT T Indicates positions of interrogation Data collection and image analysis Collect Identif Double interrogation color image beads With 2 base encoding each base is defined twice T←G←G← A Collect color image Decoding Color space sequence Identify Identify TA AC AA GA bead color beads GC CA TC Possible dinucleotides CG GT GG AG Glass slide Base zero. ATXTG TT CT AT TG GG GA Decoded sequence Color space for Y IY this sequence AT G GA Base space sequence
Applied Biosystems SOLiDTM Sequencer
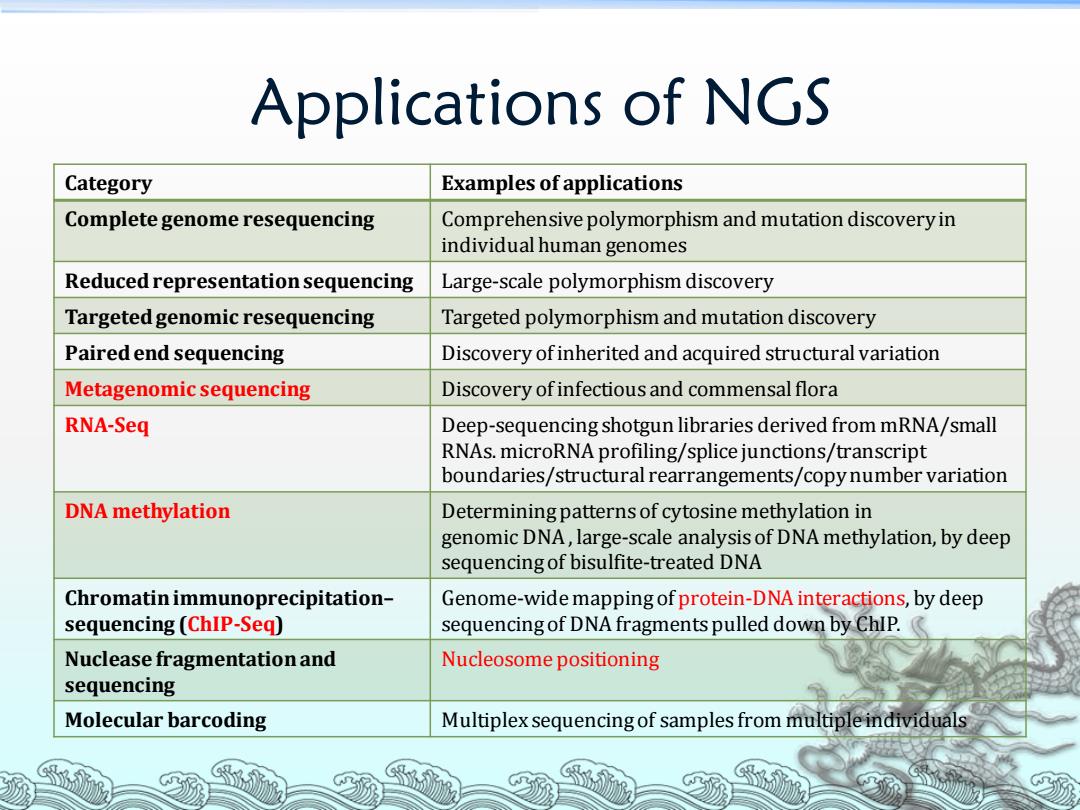
Applications of NGS Category Examples of applications Complete genome resequencing Comprehensive polymorphism and mutation discovery in individual human genomes Reduced representation sequencing Large-scale polymorphism discovery Targeted genomic resequencing Targeted polymorphism and mutation discovery Paired end sequencing Discovery of inherited and acquired structural variation Metagenomic sequencing Discovery ofinfectious and commensal flora RNA-Seq Deep-sequencing shotgun libraries derived from mRNA/small RNAs.microRNA profiling/splice junctions/transcript boundaries/structural rearrangements/copy number variation DNA methylation Determining patterns of cytosine methylation in genomic DNA,large-scale analysis of DNA methylation,by deep sequencing of bisulfite-treated DNA Chromatin immunoprecipitation- Genome-wide mapping of protein-DNA interactions,by deep sequencing(ChIP-Seq) sequencing of DNA fragments pulled down by ChIP. Nuclease fragmentation and Nucleosome positioning sequencing Molecular barcoding Multiplex sequencing of samples from multiple individuals
Applications of NGS Category Examples of applications Complete genome resequencing Comprehensive polymorphism and mutation discovery in individual human genomes Reduced representation sequencing Large-scale polymorphism discovery Targeted genomic resequencing Targeted polymorphism and mutation discovery Paired end sequencing Discovery of inherited and acquired structural variation Metagenomic sequencing Discovery of infectious and commensal flora RNA-Seq Deep-sequencing shotgun libraries derived from mRNA/small RNAs. microRNA profiling/splice junctions/transcript boundaries/structural rearrangements/copy number variation DNA methylation Determining patterns of cytosine methylation in genomic DNA , large-scale analysis of DNA methylation, by deep sequencing of bisulfite-treated DNA Chromatin immunoprecipitation– sequencing (ChIP-Seq) Genome-wide mapping of protein-DNA interactions, by deep sequencing of DNA fragments pulled down by ChIP. Nuclease fragmentation and sequencing Nucleosome positioning Molecular barcoding Multiplex sequencing of samples from multiple individuals

We are interested in .. mRNA/small RNA analyses (RNA-Seq) Analysis of mRNA targets of miRNA/siRNA Epigenetic analyses (ChIP-Seq)
We are interested in … mRNA/small RNA analyses (RNA-Seq) Analysis of mRNA targets of miRNA/siRNA Epigenetic analyses (ChIP-Seq)