
Almost Universal Code Genetic code is NOT strictly universal Certain eukaryotic nuclei and mitochondria along with at least one bacterium Codons cause termination in standard genetic code can code for amino acids Trp,Glu - Mitochondrial genomes and nuclei of at least one yeast have sense of codon changed from one amino acid to another Deviant codes are still closely related to standard one from which they evolved Genetic code a frozen accident or the product of evolution -Ability to cope with mutations evolution 18-16
18-16 Almost Universal Code • Genetic code is NOT strictly universal • Certain eukaryotic nuclei and mitochondria along with at least one bacterium – Codons cause termination in standard genetic code can code for amino acids Trp, Glu – Mitochondrial genomes and nuclei of at least one yeast have sense of codon changed from one amino acid to another • Deviant codes are still closely related to standard one from which they evolved • Genetic code a frozen accident or the product of evolution – Ability to cope with mutations evolution
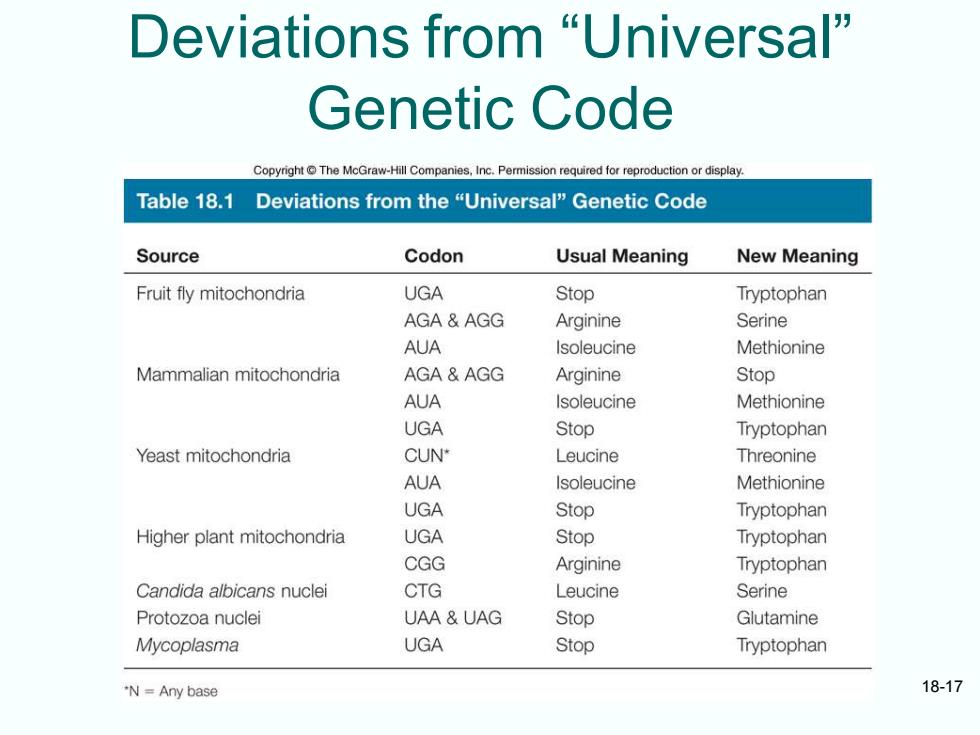
Deviations from "Universal" Genetic Code Copyright The McGraw-Hill Companies,Inc.Permission required for reproduction or display. Table 18.1 Deviations from the "Universal"Genetic Code Source Codon Usual Meaning New Meaning Fruit fly mitochondria UGA Stop Tryptophan AGA AGG Arginine Serine AUA Isoleucine Methionine Mammalian mitochondria AGA&AGG Arginine Stop AUA Isoleucine Methionine UGA Stop Tryptophan Yeast mitochondria CUN Leucine Threonine AUA Isoleucine Methionine UGA Stop Tryptophan Higher plant mitochondria UGA Stop Tryptophan CGG Arginine Tryptophan Candida albicans nuclei CTG Leucine Serine Protozoa nuclei UAA UAG Stop Glutamine Mycoplasma UGA Stop Tryptophan "N=Any base 18-17
18-17 Deviations from “Universal” Genetic Code
18.3 The Elongation Mechanism Elongation takes place in three steps: 1.EF-Tu with GTP binds aminoacyl-tRNA to the ribosomal A site 2.Peptidyl transferase forms a peptide bond between peptide in P site and newly arrived aminoacyl-tRNA in the A site Lengthens peptide by one amino acid and shifts it to the A site 3. EF-G with GTP translocates the growing peptidyl-tRNA with its mRNA codon to the P site 18-18
18-18 18.3 The Elongation Mechanism Elongation takes place in three steps: 1. EF-Tu with GTP binds aminoacyl-tRNA to the ribosomal A site 2. Peptidyl transferase forms a peptide bond between peptide in P site and newly arrived aminoacyl-tRNA in the A site Lengthens peptide by one amino acid and shifts it to the A site 3. EF-G with GTP translocates the growing peptidyl-tRNA with its mRNA codon to the P site

Elongation in Translation Copyright The McGraw-Hill Companies,Inc. ission required for reproduction or display Round 1(a) Round 1(b) EF-Tu Peptidyl transferase GTP 123 a3 Round 2(a) Round 2(b) EF-TU Peptidyl transferase GTP 2 Round 1(c) 2 EF-G GTP 12 Round 2(c) EF-G GTP 18-19
18-19 Elongation in Translation

A Three-Site Model of the Ribosome 。Puromycin Resembles an aminoacyl-tRNA -Can bind to the A site Couple with the peptide in the P site -Release it as peptidyl puromycin If peptidyl-tRNA is in the A site,puromycin will not bind to ribosome,peptide will not be released Two sites are defined on the ribosome: -Puromycin-reactive site (P) -Puromycin unreactive site(A) 3rd site (E)for deacylated tRNA bind to E site as exits ribosome 18-20
18-20 A Three-Site Model of the Ribosome • Puromycin – Resembles an aminoacyl-tRNA – Can bind to the A site – Couple with the peptide in the P site – Release it as peptidyl puromycin • If peptidyl-tRNA is in the A site, puromycin will not bind to ribosome, peptide will not be released • Two sites are defined on the ribosome: – Puromycin-reactive site (P) – Puromycin unreactive site (A) • 3 rd site (E) for deacylated tRNA bind to E site as exits ribosome