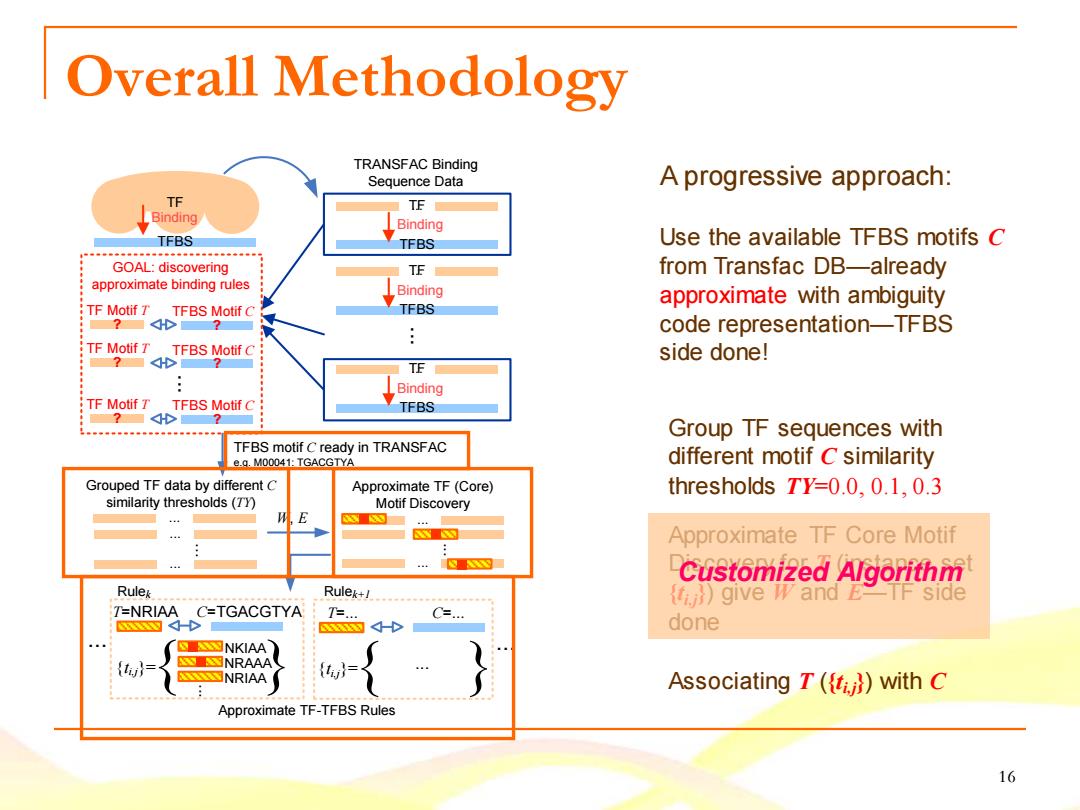
Overall Methodology TRANSFAC Binding Sequence Data A progressive approach: TF TE Binding Binding TFBS TFBS Use the available TFBS motifs C GOAL:discovering TE from Transfac DB-already approximate binding rules Binding approximate with ambiguity TF Motif T TFBS Motif C TFBS code representation-TFBS :TF Motif T TFBS Motif C side done! ?■dH■ 7 TE : Binding TF Motif T TFBS Motif C TFBS Group TF sequences with TFBS motif C ready in TRANSFAC e.g.M00041:TGACGTYA different motif C similarity Grouped TF data by different C Approximate TF(Core) thresholds TY=0.0.0.1,0.3 similarity thresholds(7Y Motif Discovery 4 E Approximate TF Core Motif Customized Algorithm Rulek Rulek+1 give W and E TF side T-NRIAA C=TGACGTYA T=. C=. done SS图NKIAA SSESNRAAA {l= Associating T((fi with C Approximate TF-TFBS Rules 16
16 Overall Methodology TF TFBS T..F. TFBS Binding TFBS T..F. Binding TFBS T..F. Binding ... GOAL: discovering approximate binding rules ... TRANSFAC Binding Sequence Data TFBS motif C ready in TRANSFAC e.g. M00041: TGACGTYA Grouped TF data by different C similarity thresholds (TY) ... ... ... ... Approximate TF (Core) Motif Discovery Binding TF Motif T TFBS Motif C ? ? TF Motif T TFBS Motif C ? ? TF Motif T TFBS Motif C ? ? Approximate TF-TFBS Rules W, E ... T=NRIAA C=TGACGTYA {ti,j}= NKIAA NRIAA NRAAA .. { . } T=... C=... {ti,j}= { ... } ... ... ... ... Rulek Rulek+1 ... Use the available TFBS motifs C from Transfac DB—already approximate with ambiguity code representation—TFBS side done! Group TF sequences with different motif C similarity thresholds TY=0.0, 0.1, 0.3 Approximate TF Core Motif Discovery for T (instance set {ti,j}) give W and E—TF side done A progressive approach: Associating T ({ti,j}) with C Customized Algorithm
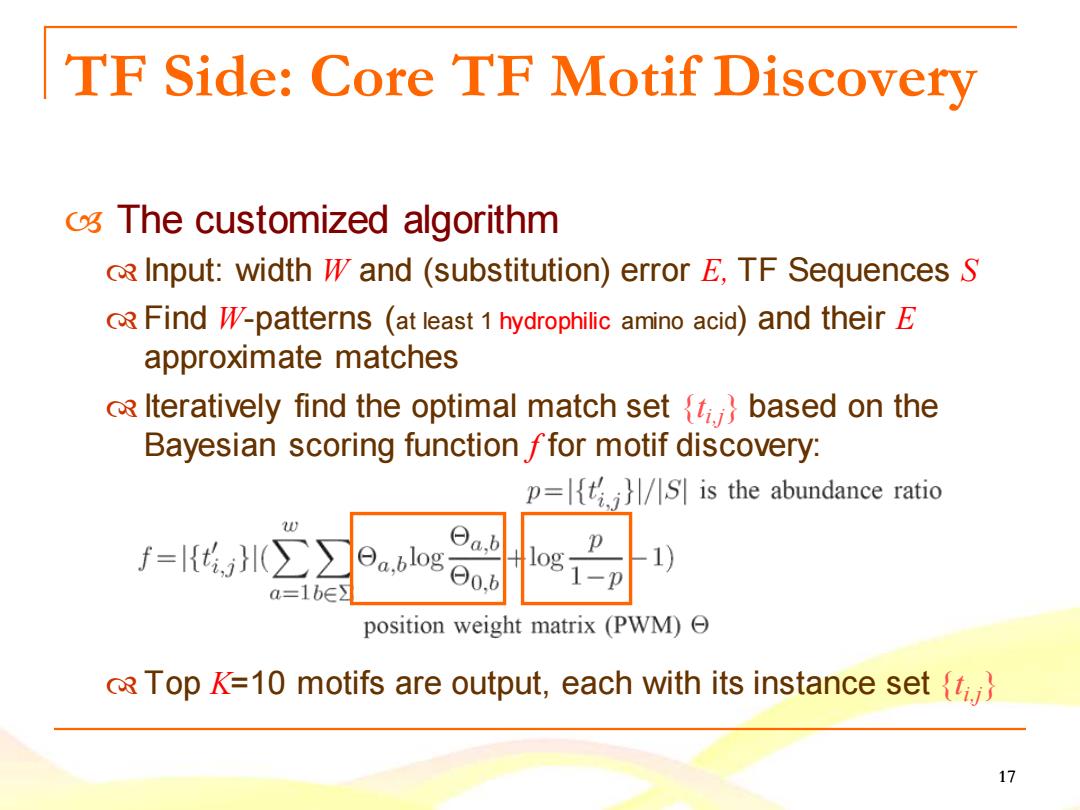
TF Side:Core TF Motif Discovery o The customized algorithm Input:width W and (substitution)error E,TF Sequences S RFind WV-patterns (at least 1 hydrophilic amino acid)and their E approximate matches o Iteratively find the optimal match set (t.}based on the Bayesian scoring function ffor motif discovery: p=S is the abundance ratio f={H(∑ ab a=1be】 日a,blo80.b position weight matrix(PWM))Θ RTop K=10 motifs are output,each with its instance set} 17
17 TF Side: Core TF Motif Discovery The customized algorithm Input: width W and (substitution) error E, TF Sequences S Find W-patterns (at least 1 hydrophilic amino acid) and their E approximate matches Iteratively find the optimal match set {ti,j} based on the Bayesian scoring function f for motif discovery: Top K=10 motifs are output, each with its instance set {ti,j}
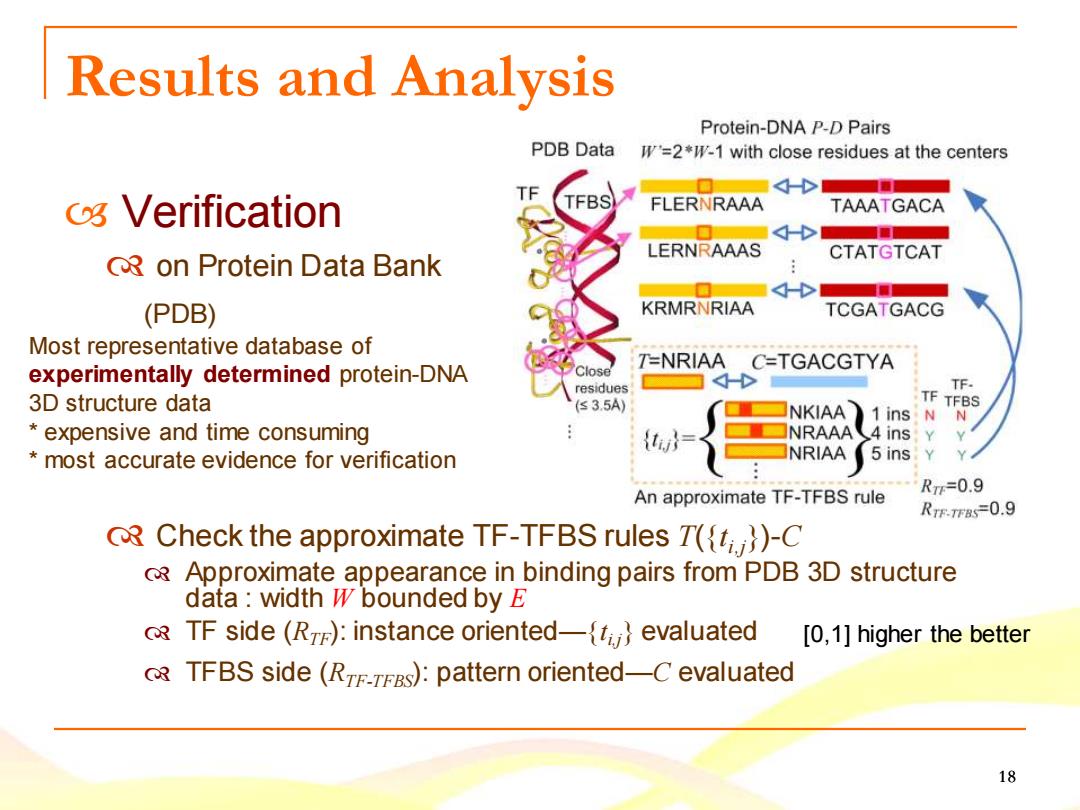
Results and Analysis Protein-DNA P-D Pairs PDB Data =2*1-1 with close residues at the centers TF 4 os Verification FLERNRAAA TAAATGACA ■ LERNRAAAS CTATGTCAT QRon Protein Data Bank (PDB) KRMRNRIAA TCGATGACG Most representative database of T-NRIAA C-TGACGTYA experimentally determined protein-DNA residues TF. 3D structure data (≤3.5A) TF NKIAA TFBS 1 ins:N expensive and time consuming {twd NRAAA4 ins Y most accurate evidence for verification NRIAA 5 ins:Y An approximate TF-TFBS rule Rm=0.9 RTE.TFBS-0.9 R Check the approximate TF-TFBS rules T((t))-C Approximate appearance in binding pairs from PDB 3D structure data:width W bounded by E 8 TF side(R):instance oriented-ti}evaluated [0,1]higher the better TFBS side(RTETES):pattern oriented-C evaluated 18
18 Verification on Protein Data Bank (PDB) Check the approximate TF-TFBS rules T({t i,j})-C Approximate appearance in binding pairs from PDB 3D structure data : width W bounded by E TF side (RTF): instance oriented—{t i,j} evaluated TFBS side (RTF-TFBS): pattern oriented—C evaluated [0,1] higher the better Results and Analysis Most representative database of experimentally determined protein-DNA 3D structure data * expensive and time consuming * most accurate evidence for verification
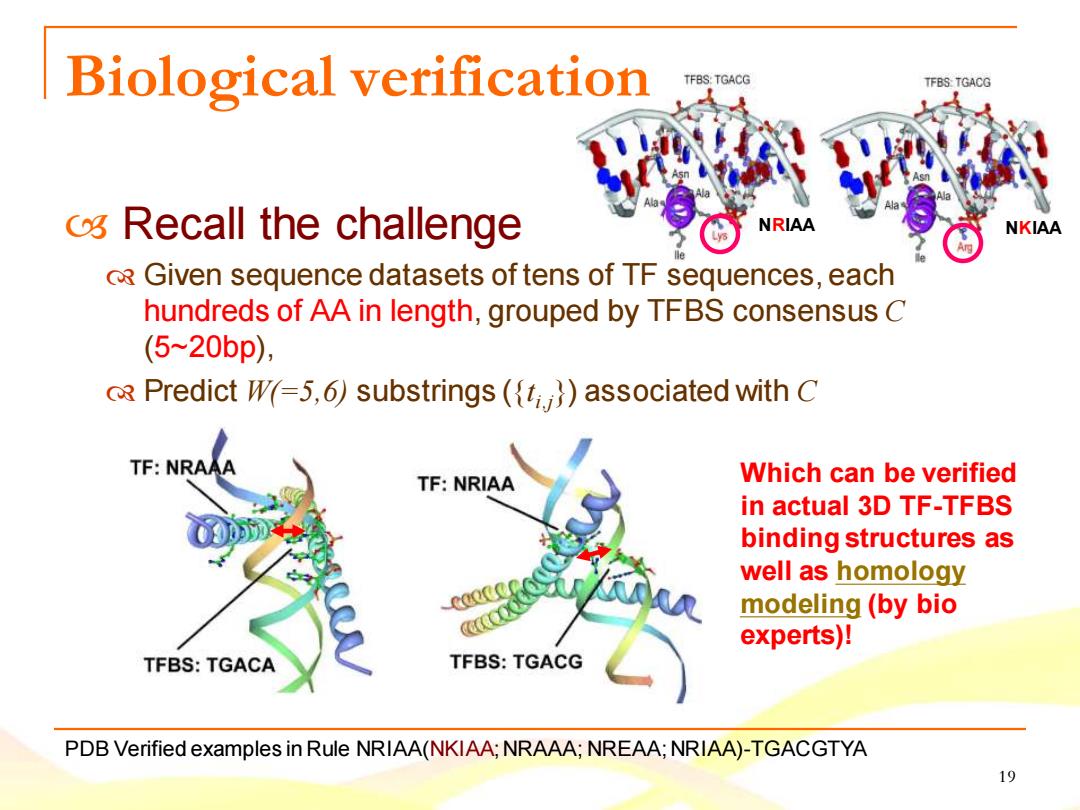
Biological verification TFBS:TGACG TFBS:TGACG os Recall the challenge NRIAA NKIAA R Given sequence datasets of tens of TF sequences,each hundreds of AA in length,grouped by TFBS consensus C (5~20bp), R Predict W=5,6)substrings(()associated with C TF:NRAAA TF:NRIAA Which can be verified in actual 3D TF-TFBS binding structures as well as homology modeling (by bio experts)! TFBS:TGACA TFBS:TGACG PDB Verified examples in Rule NRIAA(NKIAA;NRAAA;NREAA:NRIAA)-TGACGTYA 19
19 Biological verification Recall the challenge Given sequence datasets of tens of TF sequences, each hundreds of AA in length, grouped by TFBS consensus C (5~20bp), Predict W(=5,6) substrings ({t i,j}) associated with C PDB Verified examples in Rule NRIAA(NKIAA; NRAAA; NREAA; NRIAA)-TGACGTYA Which can be verified in actual 3D TF-TFBS binding structures as well as homology modeling (by bio experts)! NRIAA NKIAA
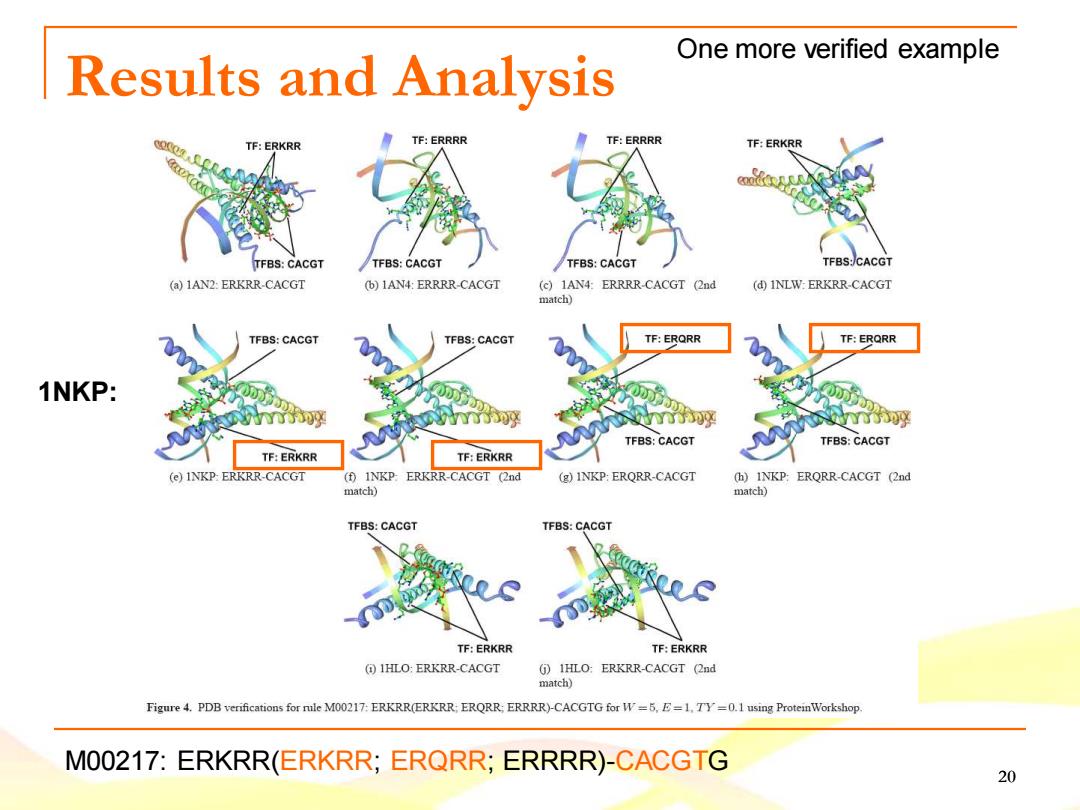
One more verified example Results and Analysis TF:ERKRR TF:ERRRR TF:ERRRR TF:ERKRR TFBS:CACGT TFBS:CACGT /TFBS:CACGT TFBS:/CACGT (a)1AN2:ERKRR-CACGT (b)1AN4:ERRRR-CACGT (c)1AN4:ERRRR-CACGT (2nd (d)INLW:ERKRR-CACGT match) TFBS:CACGT TFBS:CACGT TF:ERQRR TF:ERQRR 1NKP: y TFBS:CACGT TFBS:CACGT TF:ERKRR TF:ERKRR (e)INKP:ERKRR-CACGT (f)INKP:ERKRR-CACGT (2nd (g)INKP:ERQRR-CACGT (h)INKP:ERQRR-CACGT (2nd match) match) TFBS:CACGT TFBS:CACGT TF:ERKRR TF:ERKRR (IHLO:ERKRR-CACGT (j)IHLO:ERKRR-CACGT (2nd match) Figure 4.PDB verifications for rule M00217:ERKRR(ERKRR:ERQRR;ERRRR)-CACGTG for W=5,E=1.TY=0.1 using ProteinWorkshop. M00217:ERKRR(ERKRR:ERQRR;ERRRR)-CACGTG 20
20 Results and Analysis M00217: ERKRR(ERKRR; ERQRR; ERRRR)-CACGTG 1NKP: One more verified example