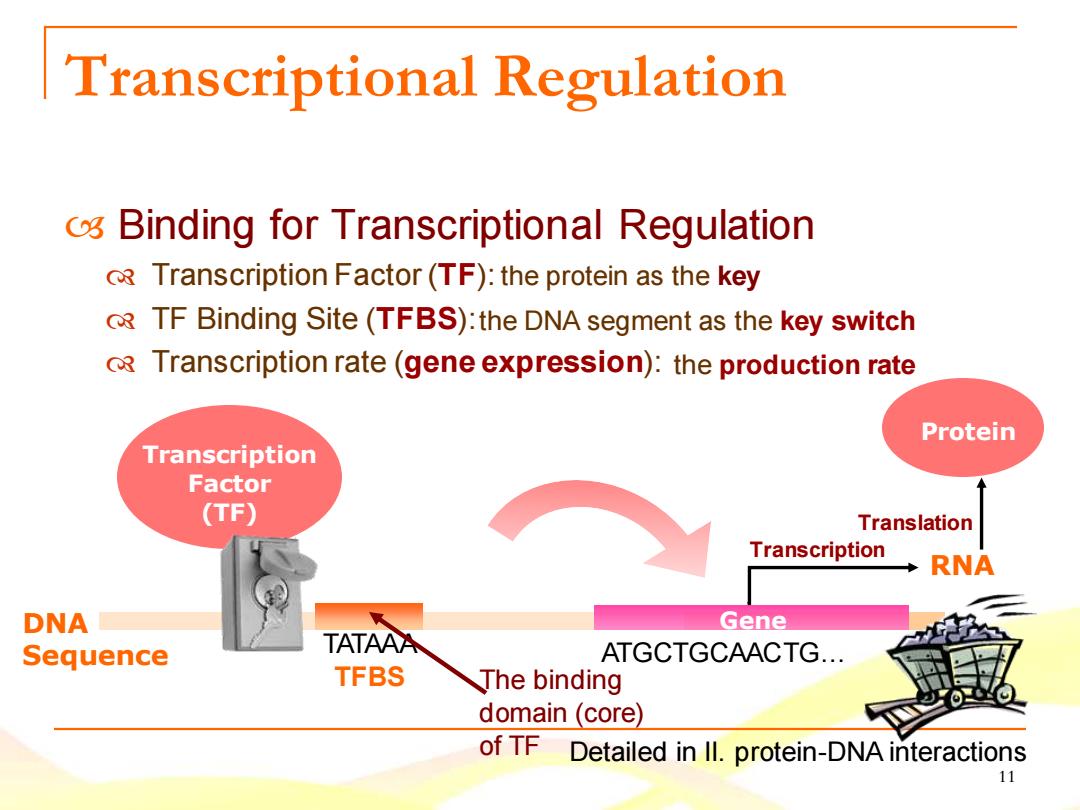
Transcriptional Regulation o Binding for Transcriptional Regulation R Transcription Factor(TF):the protein as the key R TF Binding Site(TFBS):the DNA segment as the key switch RTranscription rate (gene expression):the production rate Protein Transcription Factor (TF) Translation Transcription RNA DNA Gene Sequence TATAAA ATGCTGCAACTG... TFBS The binding domain (core) of TF Detailed in ll.protein-DNA interactions 11
11 Transcriptional Regulation Binding for Transcriptional Regulation Transcription Factor (TF): TF Binding Site (TFBS): Transcription rate (gene expression): Transcription Translation Gene RNA DNA Sequence Transcription Factor (TF) Protein TATAAA TFBS ATGCTGCAACTG… The binding domain (core) of TF the protein as the key the DNA segment as the key switch the production rate Detailed in II. protein-DNA interactions
II.Protein-DNA Interactions o Introduction o Approximate TF-TFBS rule discovery o Results and Analysis o Discussion Tak-Ming Chan.Ka-Chun Wong,Kin-Hong Lee,Man-Hon Wong,Chi-Kong Lau,Stephen Kwok-Wing Tsui,Kwong-Sak Leung,Discovering Approximate Associated Sequence Pattems for Protein-DNA Interactions.Bioinformatics,2011,27(4),pp.471-478 12
12 II. Protein-DNA Interactions Introduction Approximate TF-TFBS rule discovery Results and Analysis Discussion Tak-Ming Chan, Ka-Chun Wong, Kin-Hong Lee, Man-Hon Wong, Chi-Kong Lau, Stephen Kwok-Wing Tsui, Kwong-Sak Leung, Discovering Approximate Associated Sequence Patterns for Protein-DNA Interactions. Bioinformatics, 2011, 27(4), pp. 471-478
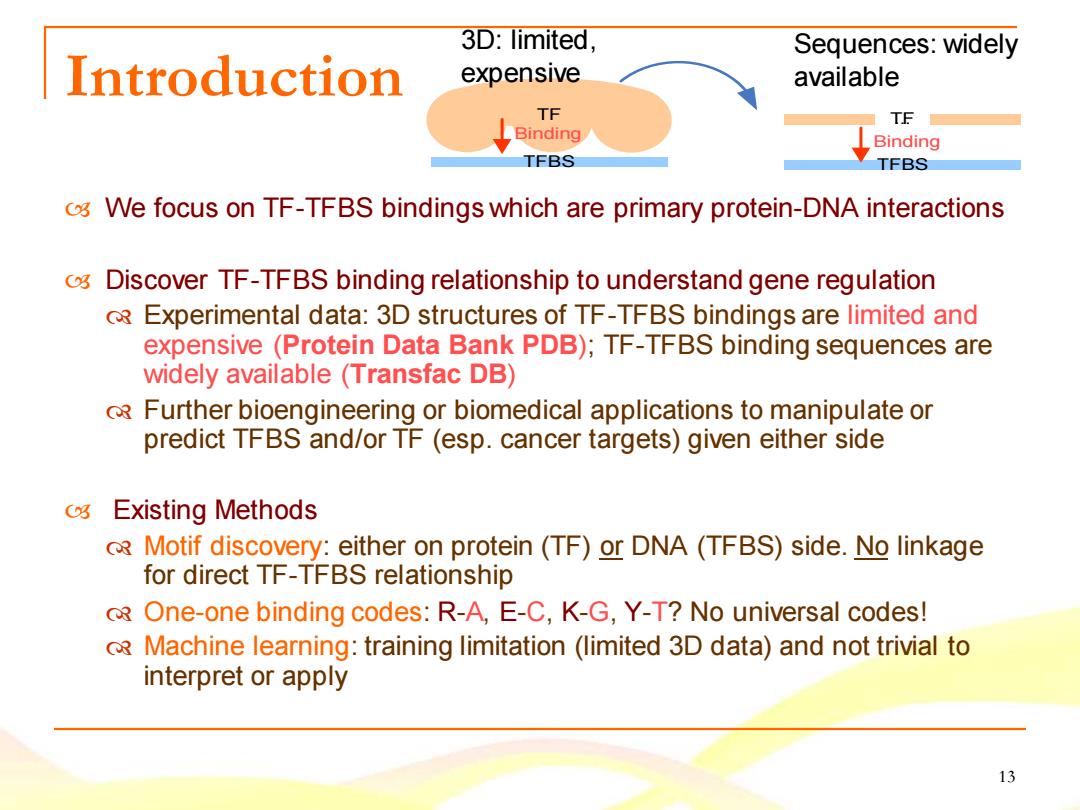
3D:limited, Sequences:widely Introduction expensive available TF TF Binding Binding TFBS TFBS o We focus on TF-TFBS bindings which are primary protein-DNA interactions o Discover TF-TFBS binding relationship to understand gene regulation RExperimental data:3D structures of TF-TFBS bindings are limited and expensive (Protein Data Bank PDB);TF-TFBS binding sequences are widely available (Transfac DB) Further bioengineering or biomedical applications to manipulate or predict TFBS and/or TF (esp.cancer targets)given either side C3 Existing Methods R Motif discovery:either on protein(TF)or DNA (TFBS)side.No linkage for direct TF-TFBS relationship 3 One-one binding codes:R-A,E-C,K-G,Y-T?No universal codes! Machine learning:training limitation (limited 3D data)and not trivial to interpret or apply 13
13 Introduction We focus on TF-TFBS bindings which are primary protein-DNA interactions Discover TF-TFBS binding relationship to understand gene regulation Experimental data: 3D structures of TF-TFBS bindings are limited and expensive (Protein Data Bank PDB); TF-TFBS binding sequences are widely available (Transfac DB) Further bioengineering or biomedical applications to manipulate or predict TFBS and/or TF (esp. cancer targets) given either side Existing Methods Motif discovery: either on protein (TF) or DNA (TFBS) side. No linkage for direct TF-TFBS relationship One-one binding codes: R-A, E-C, K-G, Y-T? No universal codes! Machine learning: training limitation (limited 3D data) and not trivial to interpret or apply TF TFBS T..F. TFBS Binding Binding Sequences: widely available 3D: limited, expensive
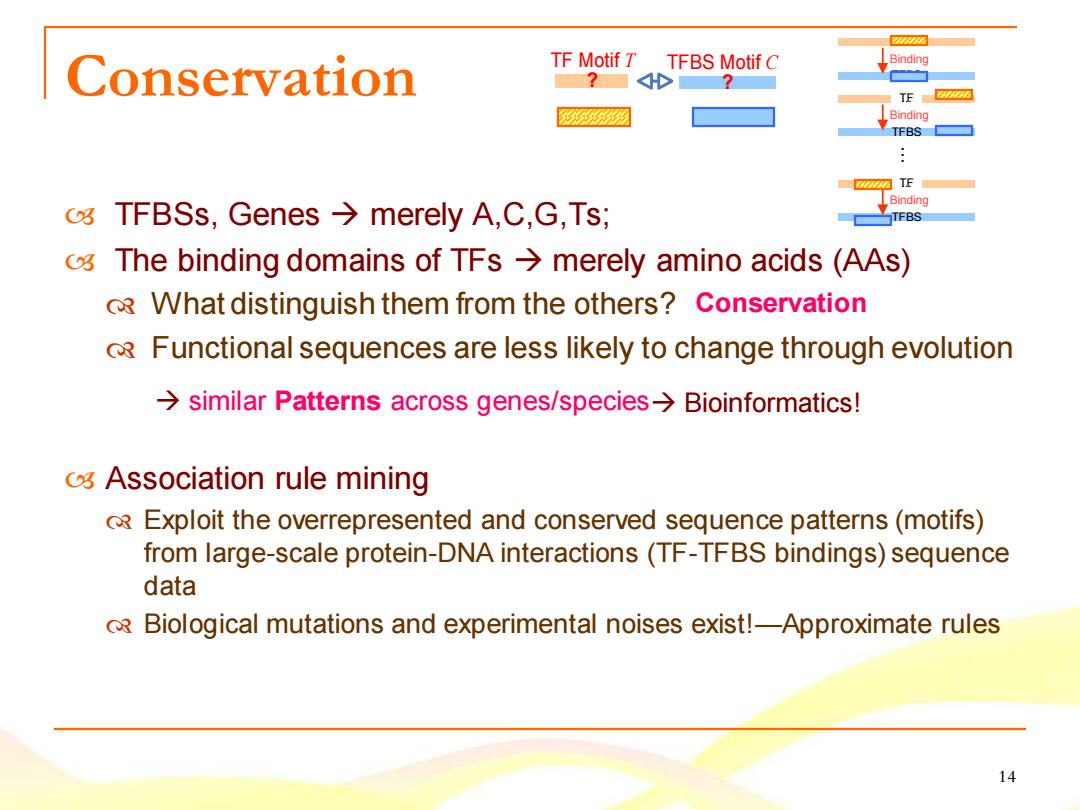
Conservation TF Motif T TFBS Motif C Binding 2 TE Binding TFBS TE Binding o TFBSs,Genes merely A,C,G,Ts; TFBS 0 The binding domains of TFs>merely amino acids(AAs) RWhat distinguish them from the others?Conservation oR Functional sequences are less likely to change through evolution similar Patterns across genes/species>Bioinformatics! o Association rule mining oRExploit the overrepresented and conserved sequence patterns(motifs) from large-scale protein-DNA interactions(TF-TFBS bindings)sequence data Biological mutations and experimental noises exist!-Approximate rules 14
14 TF Motif T TFBS Motif C Conservation ? ? TFBSs, Genes → merely A,C,G,Ts; The binding domains of TFs → merely amino acids (AAs) What distinguish them from the others? Functional sequences are less likely to change through evolution Association rule mining Exploit the overrepresented and conserved sequence patterns (motifs) from large-scale protein-DNA interactions (TF-TFBS bindings) sequence data Biological mutations and experimental noises exist!—Approximate rules Conservation → similar Patterns across genes/species→ Bioinformatics! TFBS T..F. Binding TFBS T..F. Binding TFBS T..F. Binding
Motivations GOAL:discovering approximate binding rules TF Motif T TFBS Motif C o Problem Introduction R Input:given a set of TF-TFBS binding sequences (TF:hundreds of AAs:TFBS:tens of bps depending on experiment resolution),discover the associated patterns of width W(potential interaction cores within binding distance) R Associated TF-TFBS binding sequence patterns(TF-TFBS rules) -given binding sequence data (Transfac)ONLY,predict short TF-TFBS pairs verifiable in real 3D structures of protein-DNA interactions (PDB)! o Previous method:exact Association Rule Mining based on exact counts (supports) oR Prohibited for sequence variations common in reality Simple counts can happen by chance(no elaborate modeling) Motivations:Approximation is critical! Small errors should be allowed! Model "overrepresented"biologically(probabilistic model VS counts/supports)! Kwong-Sak Leung,Ka-ChunWong.Tak-Ming Chan,Man-Hon Wong.Kin-Hong Lee,Chi-Kong Lau,Stephen Kwok-Wing Tsui,Discovering Protein-DNA Binding Sequence Patterns Using Association Rule Mining.Nucleic Acids Research.2010,38(19),pp.6324-6337 15
15 Motivations Problem Introduction Input: given a set of TF-TFBS binding sequences (TF: hundreds of AAs; TFBS: tens of bps depending on experiment resolution), discover the associated patterns of width w (potential interaction cores within binding distance) Associated TF-TFBS binding sequence patterns (TF-TFBS rules) —given binding sequence data (Transfac) ONLY, predict short TF-TFBS pairs verifiable in real 3D structures of protein-DNA interactions (PDB)! Previous method: exact Association Rule Mining based on exact counts (supports) Prohibited for sequence variations common in reality Simple counts can happen by chance (no elaborate modeling) Motivations: Approximation is critical! Small errors should be allowed! Model “overrepresented” biologically (probabilistic model VS counts/supports)! GOAL: discovering approximate binding rules TF Motif T TFBS Motif C ? ? Kwong-Sak Leung, Ka-ChunWong, Tak-Ming Chan, Man-Hon Wong, Kin-Hong Lee, Chi-Kong Lau, Stephen Kwok-Wing Tsui, Discovering Protein-DNA Binding Sequence Patterns Using Association Rule Mining. Nucleic Acids Research. 2010, 38(19), pp. 6324-6337