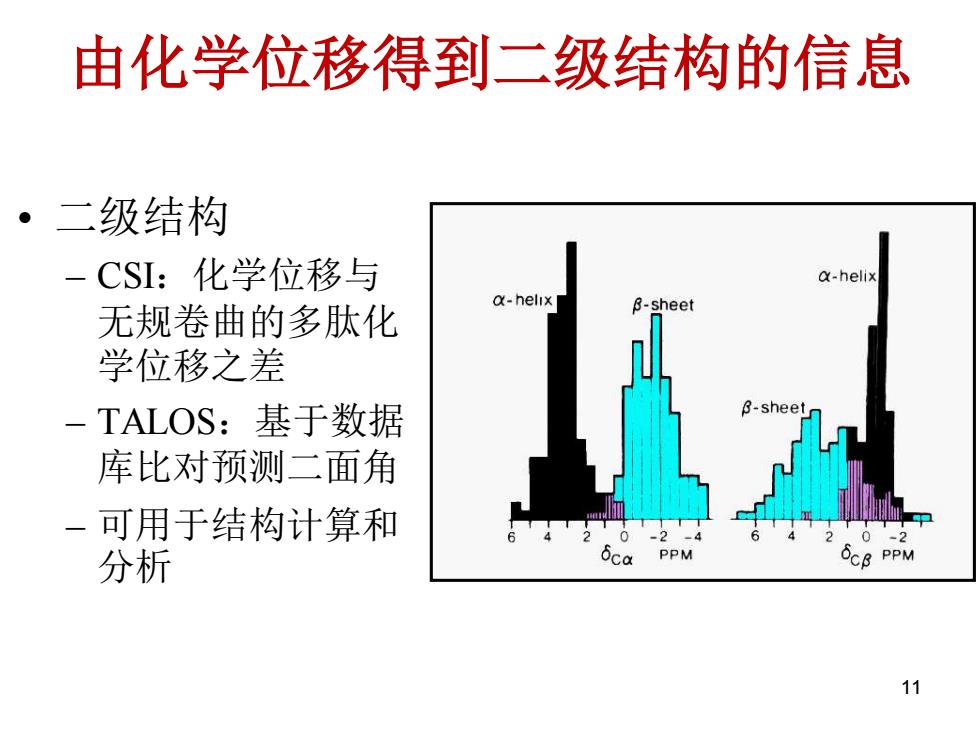
由化学位移得到二级结构的信息 。二级结构 CSI:化学位移与 a-helix a-helix -shee 无规卷曲的多肽化 学位移之差 -TALOS:基于数据 B-sheet 库比对预测二面角 可用于结构计算和 22 分析 PPM OCB PPM 11
由化学位移得到二级结构的信息 • 二级结构 – CSI:化学位移与 无规卷曲的多肽化 学位移之差 – TALOS:基于数据 库比对预测二面角 – 可用于结构计算和 分析 11
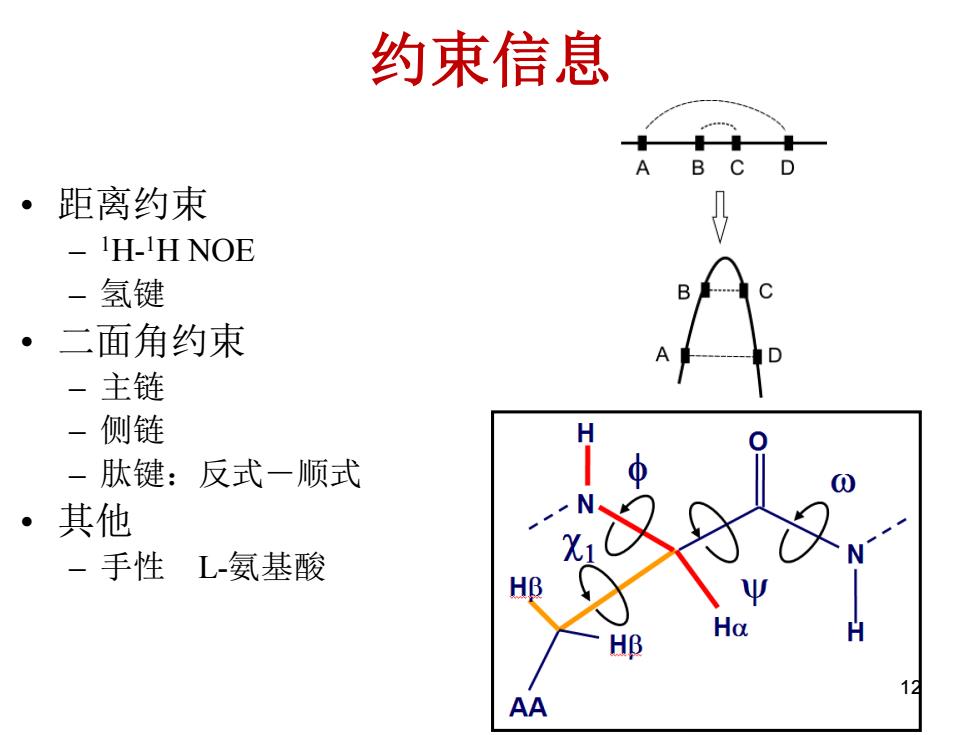
约束信息 。 距离约束 -H-H NOE - 氢键 B ·二面角约束 一主链 一侧链 -肽键: 反式一顺式 ·其他 -手性 L-氨基酸 HB Ho 1 AA
约束信息 • 距离约束 – 1H- 1H NOE – 氢键 • 二面角约束 – 主链 – 侧链 – 肽键:反式-顺式 • 其他 – 手性 L-氨基酸 12
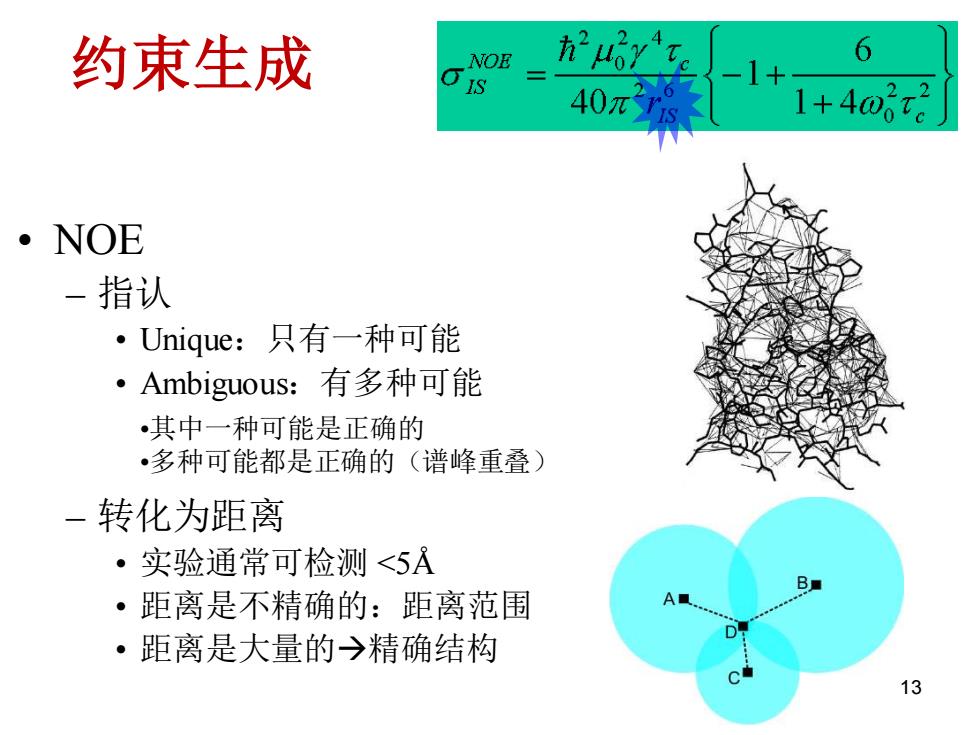
约束生成 NOE 6 40π 2 1+40。 ·NOE 一指认 ·Unique:只有一种可能 ·Ambiguous:有多种可能 •其中一种可能是正确的 •多种可能都是正确的(谱峰重叠) -转化为距离 ·实验通常可检测<5A B ·距离是不精确的:距离范围 A D ·距离是大量的→精确结构 C 13
约束生成 • NOE – 指认• Unique:只有一种可能 • Ambiguous:有多种可能 – 转化为距离 • 实验通常可检测 <5Å • 距离是不精确的:距离范围 • 距离是大量的→精确结构 •其中一种可能是正确的 •多种可能都是正确的(谱峰重叠) 13
NMR data 1:NOE For short mixing times NOE cross peak intensity is proportional to 1/r6 of two protons. 。NOE~1/r6ftc) For well structured areas of a macromolecule f(t)can be considered to be constant.(in practice this is assumed to be true for all parts of the molecule) -Calibration of cross peaks by using a proton pair of known local geometry (distance) -Because of multiple simplifying assumptions of the relationship between NOE and distance it is usually used only qualitatively(class NOEs in three bins:strong,medium and weak) 14
NMR data 1: NOE • For short mixing times NOE cross peak intensity is proportional to 1/r6 of two protons. • NOE ~ 1/r6 f(tc ) – For well structured areas of a macromolecule f(tc ) can be considered to be constant. (in practice this is assumed to be true for all parts of the molecule) – Calibration of cross peaks by using a proton pair of known local geometry (distance) – Because of multiple simplifying assumptions of the relationship between NOE and distance it is usually used only qualitatively (class NOEs in three bins: strong, medium and weak) 14
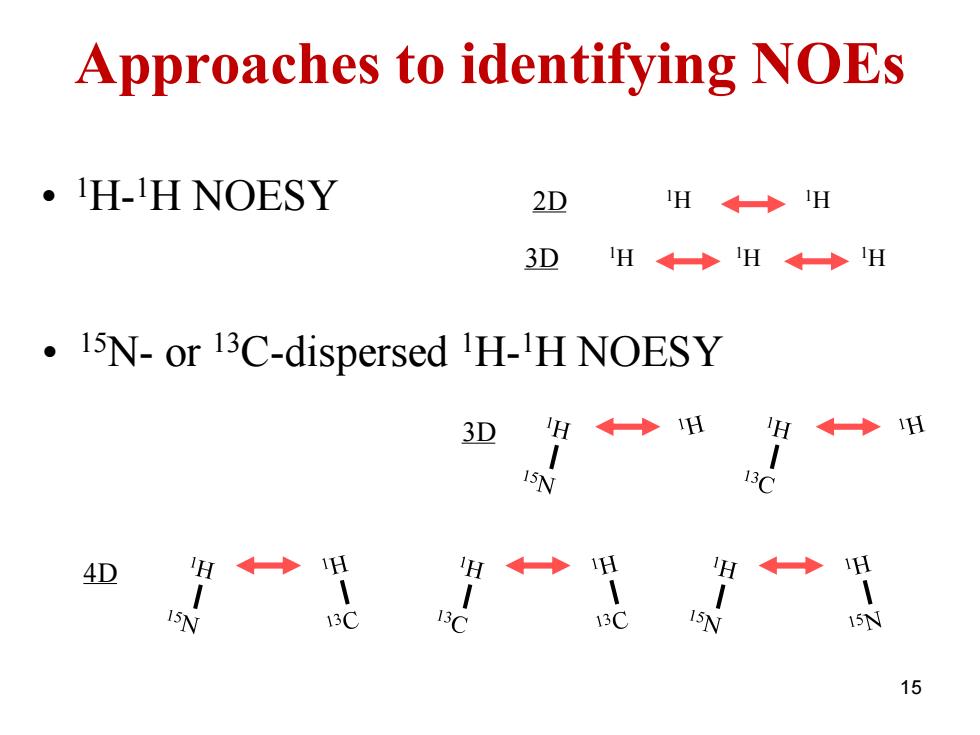
Approaches to identifying NOEs ·H-1 H NOESY 2D H←◆H 3D H←◆H←◆H 15N-or 13C-dispersed H-H NOESY 3D H ◆H H H 15N 13C 4D H ◆ H H←◆ H H◆◆ H 15N 13C 13C 13C 15N 15N 15
15 Approaches to identifying NOEs • 15N- or 13C-dispersed 1H- 1H NOESY 3D 4D 2D 1H 1H 3D 1H 1H 1H • 1H- 1H NOESY