06 Sample: nt containing 7.0 60 Elution buffer: 0 3.0 250 Fig.Scouting for optimal clution pH of a mo onal IgG,from HiTrap rProtein A FF using a pH gradicn Flow rates It is not possible to specify a single optimal flow rate in affinity hromatography because dissociation rates of ligand/target molecule interactions vary widely. For ready to use affinity media follow the manufacturer's instructions and optimize further if required: -determine the optimal flow rate to achieve efficient binding -determine the optimal flow rate for elution to maximize recovery -determine the maximum flow rate for column re-equilibration to minimize total run times To obtain sharp elution curves and maximum recovery with minimum dilution of separated molecules,use the lowest acceptable flow rate. Analysis of results and further steps The anai ofeocaindcate if the pufond to b en r purity,spe igh resolution gel filtration to remove any aggregates,or ligands that may have leach from the medium.For example,Superdex can be used to separate molecules,according to differences in size,and to transfer the sample into storage buffer,removing excess salt and other small molecules.The chromatogram will also give an indication of the homogeneity of the purified sample. Alternatively,a desalting column that gives low resolution,but high sample capacity,can be used to quickly transfer the sample into storage buffer and remove excess salt(see page 133). Equipment selection Appendix 2 provides a guide to the selection of purification systems 21
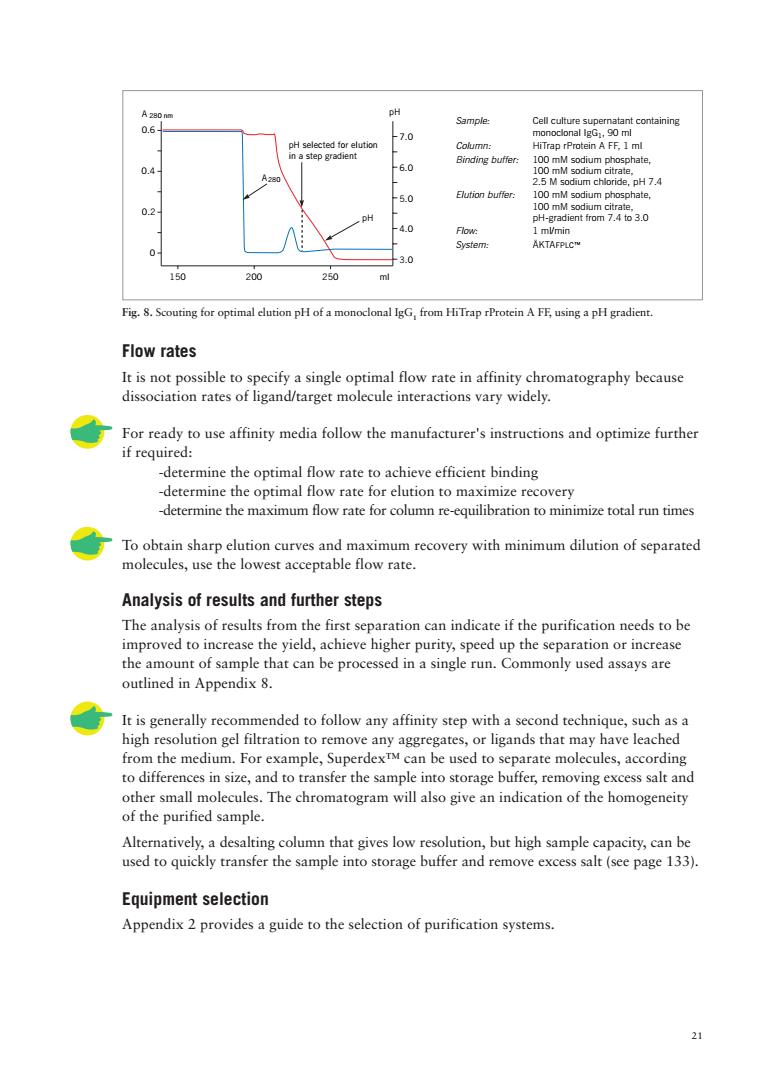
21 Fig. 8. Scouting for optimal elution pH of a monoclonal IgG1 from HiTrap rProtein A FF, using a pH gradient. Flow rates It is not possible to specify a single optimal flow rate in affinity chromatography because dissociation rates of ligand/target molecule interactions vary widely. For ready to use affinity media follow the manufacturer's instructions and optimize further if required: -determine the optimal flow rate to achieve efficient binding -determine the optimal flow rate for elution to maximize recovery -determine the maximum flow rate for column re-equilibration to minimize total run times To obtain sharp elution curves and maximum recovery with minimum dilution of separated molecules, use the lowest acceptable flow rate. Analysis of results and further steps The analysis of results from the first separation can indicate if the purification needs to be improved to increase the yield, achieve higher purity, speed up the separation or increase the amount of sample that can be processed in a single run. Commonly used assays are outlined in Appendix 8. It is generally recommended to follow any affinity step with a second technique, such as a high resolution gel filtration to remove any aggregates, or ligands that may have leached from the medium. For example, Superdex™ can be used to separate molecules, according to differences in size, and to transfer the sample into storage buffer, removing excess salt and other small molecules. The chromatogram will also give an indication of the homogeneity of the purified sample. Alternatively, a desalting column that gives low resolution, but high sample capacity, can be used to quickly transfer the sample into storage buffer and remove excess salt (see page 133). Equipment selection Appendix 2 provides a guide to the selection of purification systems. A 280 nm 3.0 4.0 5.0 6.0 7.0 150 200 250 0 0.2 0.4 0.6 ml pH A pH selected for elution in a step gradient pH 280 Sample: Cell culture supernatant containing monoclonal IgG1, 90 ml Column: HiTrap rProtein A FF, 1 ml Binding buffer: 100 mM sodium phosphate, 100 mM sodium citrate, 2.5 M sodium chloride, pH 7.4 Elution buffer: 100 mM sodium phosphate, 100 mM sodium citrate, pH-gradient from 7.4 to 3.0 Flow: 1 ml/min System: ÄKTAFPLC™

Troubleshooting This section focuses on practical problems that may occur when running a chromatography column.The diagrams below give an indication of how a chromatogram may deviate from the ideal during affinity purification and what measures can be taken to improve the results. Target elutes as a sharp peak.Satisfactory result .If it is difficult or impossible to retain biological activity when achieving this result,either new conditions or a new ligand must be found. .If using low pH for eluti ,collect the fractions in ne Target is a broad,low peak that elutes while binding buffer is being applied Find better binding conditions. Target elutes in a broad,low peak Try different elution conditions. ·e conce of the competitor in the eution buffer. during tiontoallow collect the target protein in pulses(see second figure beneath). Note:This result may also be seen if the target protein has denatured and aggregated on the column or if there is non-specific binding. Some of the target molecule elutes as a broad,low peak while still under binding conditions ·mae utes between each sample application second figure beneath)
22 Troubleshooting This section focuses on practical problems that may occur when running a chromatography column. The diagrams below give an indication of how a chromatogram may deviate from the ideal during affinity purification and what measures can be taken to improve the results. Target elutes as a sharp peak. Satisfactory result • If it is difficult or impossible to retain biological activity when achieving this result, either new elution conditions or a new ligand must be found. • If using low pH for elution, collect the fractions in neutralization buffer (60–200 µl 1 M Tris-HCl, pH 9.0 per ml eluted fraction). Target is a broad, low peak that elutes while binding buffer is being applied • Find better binding conditions. A280 ml Binding buffer Elution buffer Eluted target Flow through (unbound material) A280 ml Binding buffer Eluted target Flow through (unbound material) A280 ml Binding buffer Elution buffer Eluted target Flow through (unbound material) A280 ml Binding buffer Elution buffer Eluted target Flow through (unbound material) A280 ml Binding buffer Elution buffer Eluted target Wait Flow through (unbound material) A280 ml Binding buffer Elution buffer Eluted target Flow through (unbound material) Target elutes in a broad, low peak • Try different elution conditions. • If using competitive elution, increase the concentration of the competitor in the elution buffer. • Stop flow intermittently during elution to allow time for the target molecule to elute and so collect the target protein in pulses (see second figure beneath). Note: This result may also be seen if the target protein has denatured and aggregated on the column or if there is non-specific binding. Some of the target molecule elutes as a broad, low peak while still under binding conditions • Allow time for the sample to bind and/or apply sample in aliquots, stopping the flow for a few minutes between each sample application (see second figure beneath)

Situation Cause Remedy Sample has not been fiered properly. Clean the column,fier the sample Samole has altered durine storae er sample into Hmeter prepare reenerate thema Column is overloaded with sample. Decrease the samnle load acurred olumnsstoen2zO%ethanolwhenposib hnge or clean the Tesomtheacnd Lower yield than expected. eaegypoee Sample precipitates. atsor cengeaceneaogana hicants polaryreduing 内e e applied to thecolumn. the chromatographic step. Reorng thru ou RmPpeeoroehancdeeeeaeduring tion in the Modify the eluent to maintan stablity. nove and n or use a new colum Precipitated proteins. Bed compressed. Repack the comn, Microbial growth. th rarely occurs in columns 23
23 Situation Cause Remedy Protein does not bind Sample has not been filtered properly. Clean the column, filter the sample or elute as expected. and repeat. Sample has altered during storage. Prepare fresh samples. Sample has wrong pH or buffer Use a desalting column to transfer sample into conditions are incorrect. the correct buffer (see page 133). Solutions have wrong pH. Calibrate pH meter, prepare new solutions and try again. The column is not equilibrated Repeat or prolong the equilibration step. sufficiently in the buffer. Proteins or lipids have Clean and regenerate the column or use a precipitated on the column. new column. Column is overloaded with sample. Decrease the sample load. Microbial growth has occurred Microbial growth rarely occurs in columns in the column. during use, but, to prevent infection of packed columns, store in 20% ethanol when possible. Precipitation of protein in the Clean the column, exchange or clean the filter column filter and/ or at the top or use a new column. of the bed. Low recovery of activity, but Protein may be unstable or Determine the pH and salt stability of the normal recovery of protein. inactive in the elution buffer. protein. Collect fractions into neutralization buffer such as 1 M Tris-HCl, pH 9 (60–200 µl per fraction). Enzyme separated from Test by pooling aliquots from the fractions and co-factor or similar. repeating the assay. Lower yield than expected. Protein may have been Add protease inhibitors to the sample and degraded by proteases. buffers to prevent proteolytic digestion. Run sample through a medium such as Benzamidine 4 Fast Flow (high sub) to remove serine proteases. Adsorption to filter during Use another type of filter. sample preparation. Sample precipitates. May be caused by removal of salts or unsuitable buffer conditions. Hydrophobic proteins. Use chaotropic agents, polarity reducing agents Protein is still attached to ligand. or detergents. More activity is Different assay conditions have Use the same assay conditions for all the recovered than was been used before and after assays in the purification scheme. applied to the column. the chromatographic step. Removal of inhibitors during separation. Reduced or poor flow Presence of lipoproteins or Remove lipoproteins and aggregrates during through the column. protein aggregates. sample preparation (see Appendix 1). Protein precipitation in the Modify the eluent to maintain stability. column caused by removal of stabilizing agents during fractionation. Clogged column filter. Replace the filter or use a new column. Always filter samples and buffer before use. Clogged end-piece or Remove and clean or use a new column. adaptor or tubing. Precipitated proteins. Clean the column using recommended methods or use a new column. Bed compressed. Repack the column, if possible, or use a new column. Microbial growth. Microbial growth rarely occurs in columns during use, but, to prevent infection of packed columns, store in 20% ethanol when possible

Situation Cause Remedy Turbid sample Bubbles in the bed. Buffersnot properly de-assed. De-gas buffers thoroughly. Cracks in the bed. Large air leak in column. hgea流a药ampe Agbubteathetopothe Retall the adap taking are tor Column poorly packed. Bed packed at 24
24 Situation Cause Remedy Back pressure increases Turbid sample. Improve sample preparation (see Appendix 1). during a run or during Improve sample solubility by the addition of successive runs. ethylene glycol, detergents or organic solvents. Precipitation of protein in the column Clean using recommended methods. Exchange filter and/or at the top of the bed. or clean filter or use a new column. Include any additives that were used for initial sample solubilization in the solutions used for chromatography. Bubbles in the bed. Column packed or stored at Remove small bubbles by passing de-gassed cool temperature and then buffer upwards through the column. Take warmed up. special care if buffers are used after storage in a fridge or cold-room. Do not allow column to warm up due to sunshine or heating system. Repack column, if possible, (see Appendix 3). Buffers not properly de-gassed. De-gas buffers thoroughly. Cracks in the bed. Large air leak in column. Check all connections for leaks. Repack the column if possible (see Appendix 3). Distorted bands as sample Air bubble at the top of the Re-install the adaptor taking care to avoid air runs into the bed. column or in the inlet adaptor. bubbles. Particles in buffer or sample. Filter or centrifuge the sample. Protect buffers from dust. Clogged or damaged net in Dismantle the adaptor, clean or replace the net. upper adaptor. Keep particles out of samples and eluents. Distorted bands as Column poorly packed. Suspension too thick or too thin. Bed packed at sample passes down a temperature different from run. the bed. Bed insufficiently packed (too low packing pressure, too short equilibration). Column packed at too high pressure

Chapter 3 Purification of specific groups of molecules A group specific medium has an affinity for a group of related substances rather than for a single type of molecule.The same general ligand can be used to purify several substances (for example members of a class of enzymes)without the need to prepare a new medium for each different substance in the group.Within each group there is either structural or functional similarity.The specificity of the affinity medium derives from the selectivity of the ligand and the use of selective elution conditions. Immunoglobulins The diversity of antibody-antigen interactions has created many use s for antibodies and antibody f They are us peutic ar applic immunochemical aniques within general The ue at recom nology has greatly expanded our ability to manipulate the characteristics of the se molecules to our advantage.The potential exists to create an infinite number of combinations between immunoglobulins and immunoglobulin fragments with tags and other selected proteins. A significant advantage for the purification of antibodies and their fragments is that a great deal of information is available about the properties of the target molecule and the major contaminants,no matter whether the molecule is in its a native state or has been genetically engineered and no matter what the source material. The Antibody Purification Handbook from Amersham Biosciences presents the most effective and frequently used strategies for sample preparation and purification of the many different forms of antibodies and antibody fragments used in the laboratory The handbook also includes more detailed information on antibody structure and classification,illustrated briefly here in Figures 9 and 10. H-chain Fc-fragment Fig9.H212 structure of a typical immunoglobulin. 25
25 Chapter 3 Purification of specific groups of molecules A group specific medium has an affinity for a group of related substances rather than for a single type of molecule. The same general ligand can be used to purify several substances (for example members of a class of enzymes) without the need to prepare a new medium for each different substance in the group. Within each group there is either structural or functional similarity. The specificity of the affinity medium derives from the selectivity of the ligand and the use of selective elution conditions. Immunoglobulins The diversity of antibody-antigen interactions has created many uses for antibodies and antibody fragments. They are used for therapeutic and diagnostic applications as well as for immunochemical techniques within general research. The use of recombinant technology has greatly expanded our ability to manipulate the characteristics of these molecules to our advantage. The potential exists to create an infinite number of combinations between immunoglobulins and immunoglobulin fragments with tags and other selected proteins. A significant advantage for the purification of antibodies and their fragments is that a great deal of information is available about the properties of the target molecule and the major contaminants, no matter whether the molecule is in its a native state or has been genetically engineered and no matter what the source material. The Antibody Purification Handbook from Amersham Biosciences presents the most effective and frequently used strategies for sample preparation and purification of the many different forms of antibodies and antibody fragments used in the laboratory. The handbook also includes more detailed information on antibody structure and classification, illustrated briefly here in Figures 9 and 10. Fig. 9. H2L2 structure of a typical immunoglobulin